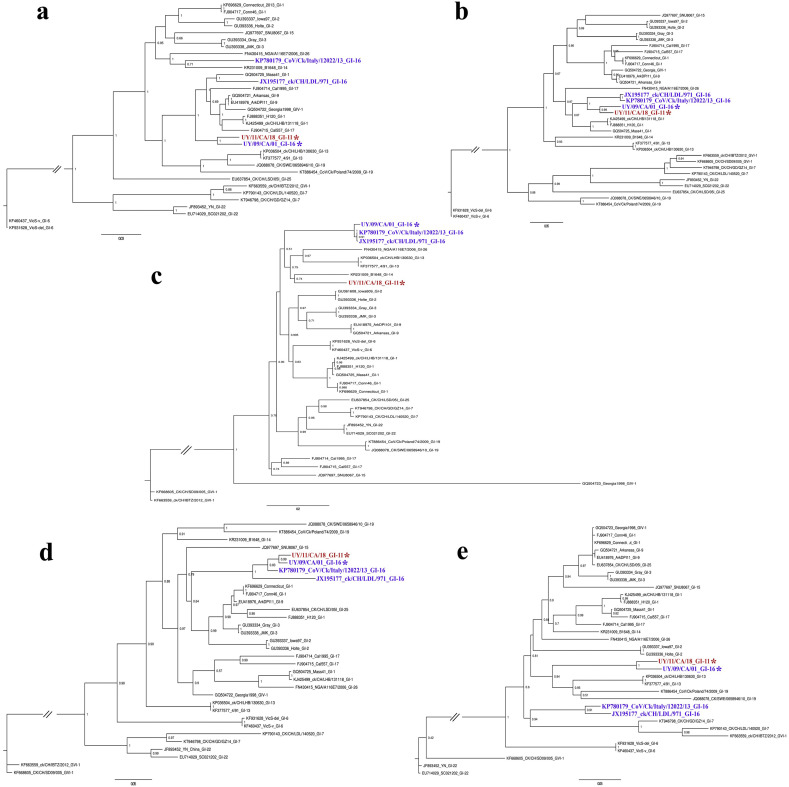
Fig. 1.
Phylogenetic trees obtained with the maximum-likelihood method and JC model (a), GTR model with gamma distribution and invariant sites (b and c), and HKY model with gamma distribution and invariant sites (d and e). Phylogenetic reconstruction was carried out using the ORF1a (a), ORF1b (b), S1 sequence (c), S2 sequence and ORFs 3a, 3b, E, M, 5a and 5b (d) and ORF N (e). Mapping uncertainties for internal nodes are shown as approximate likelihood ratio test values. Strains of GI-16 and GI-11 lineages are labeled with an asterisk. This analysis was performed using a sub-sampled non-recombinant data set with representatives of the IBV variability.