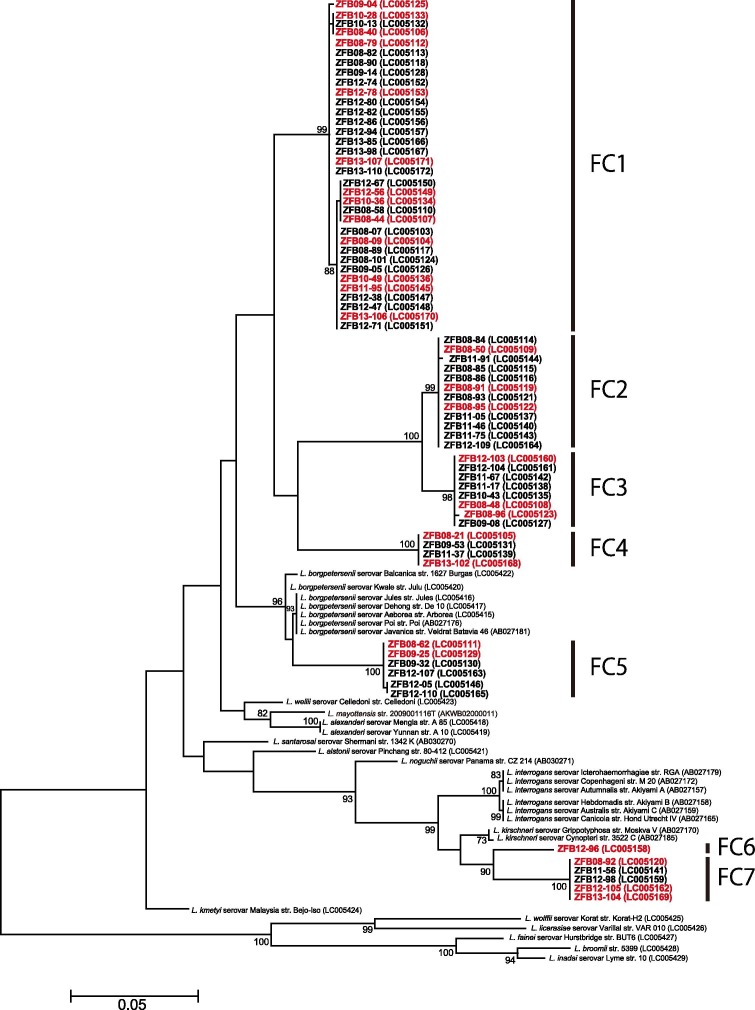
Fig. 1.
Maximum-likelihood phylogenetic tree based on the nucleotide sequences of Leptospira spp. flaB in E. helvum bats. The dendrogram was constructed with the JC69 model, and with 1,000 replications using MEGA 5.2.2 software (Tamura et al., 2011). Numbers at nodes indicate bootstrap supports >70%. The sequences determined in this study are shown in bold. The samples colored red were also used in the phylogenetic analysis of rrs (Fig. 2). GenBank accession numbers are indicated in parentheses. Scale bar indicates the number of nucleotide substitutions per site.