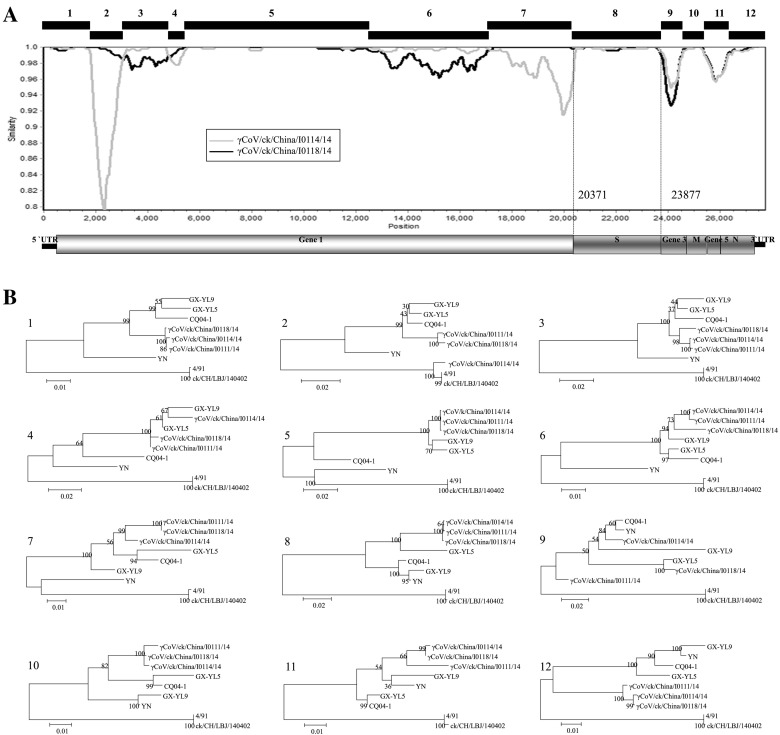
Fig. 3.
Complete genomic analysis of our three IBV isolates and the reference strains. (A) Simplot analysis between our three IBV isolates. The SimPlot was created using a 500 bp window with a 50-bp step with the γCoV/ck/China/I0111/14 isolate as the query strain. Twelve gene fragments were manually obtained based on the topologies of SimPlot analysis. (B) Phylogenetic trees were constructed using the maximum likelihood method with the general time-reversible nucleotide substitution model and bootstrap tests of 1000 replicates in the MEGA 6 program, using the 12 gene fragments (designated as 1–12) based on the above-mentioned results. Three IBV strains isolated in this study and six reference IBV strains were included. The GenBank accession numbers for these genome sequences are as follows: GX-YL9 (HQ850618), GX-YL5 (HQ848267), CQ04-1(HM245924), YN(JF893452), 4/91(KF377577) and ck/CH/LBJ/140402 (KF377577).