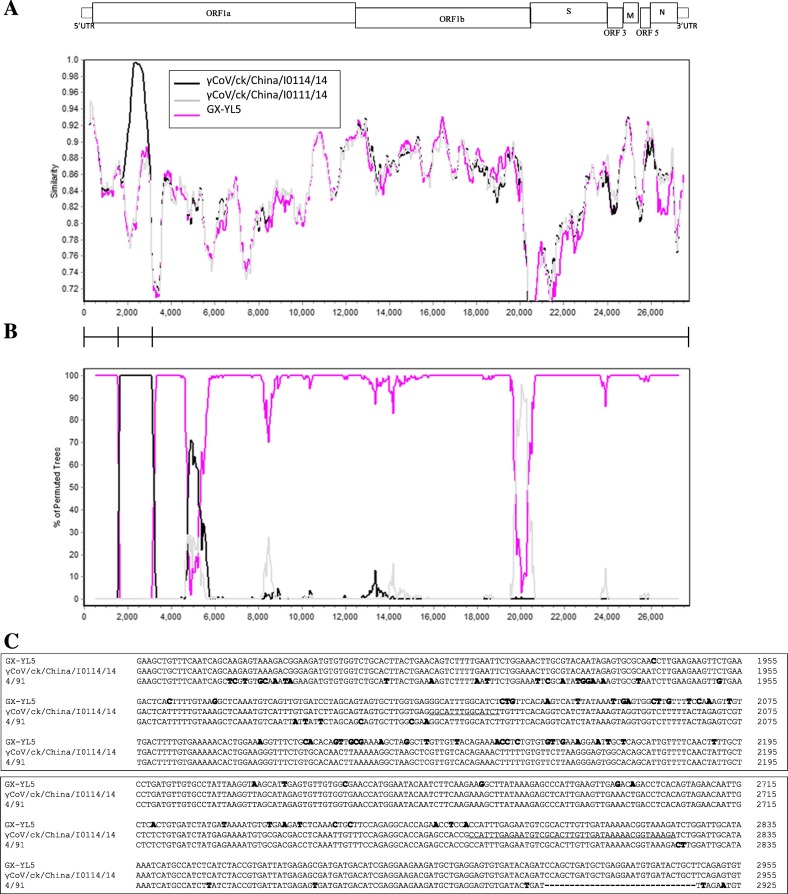
Fig. 4.
SimPlot and sequence analyses to detect recombination and estimate breakpoint within the γCoV/ck/China/I0111/14 genome. (A) Relative gene location and similarity plot. (B) Bootscan analysis showing multiple breakpoints. The 4/91 strain was used as the query sequence. The complete genomic nucleotide sequences used in the analysis were γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and GX-YL5. Both analyses were performed with the F84 distance model, with a window size of 500 bp and step size of 50 bp. (C) Multiple sequence alignment of the predicted breakpoint and flanking sequences among the strains GX-YL5, γCoV/ck/China/I0114/14 and 4/91. Numbers to the right of each alignment show the nucleotide positions in the genome of each virus. Nucleotides differing from those of the γCoV/ck/China/I0114/14 isolate are given in bold.