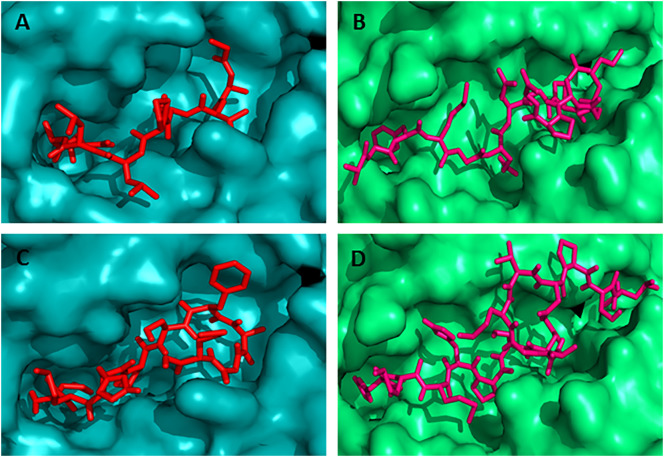
Fig. 3.
Docking simulation to predict the binding of predicted and control CD8 + and CD4 + T-cell epitopes to MHC class I (HLA-C*12:03) and MHC class II (HLA-DRB1*01:01) molecule, respectively. The colors, teal cyan and lime green indicate the surface structure of HLA-C*12:03 and HLA-DRB1*01:01, respectively. The red and hot pink colors indicate the CD8 + and CD4 + T-cell epitope, respectively (A) binding of control CD8 + T-cell epitope (GAVDPLLAL) to the binding groove of HLA-C*12:03 (affinity: − 8.4 kcal/mol) (B) represents the binding of control CD4 + T-cell epitope (AGFKGEQGPKGEPG) to the binding groove of HLA-DRB1*01:01 (affinity: − 7.3 kcal/mol) (C) shows the binding affinity of N protein CD8 + T-cell epitope “MMHPSFAGM” to the binding groove of HLA-C*12:03 (affinity: − 7.5 kcal/mol) (D) binding of N protein CD4 + T-cell epitope “HMMHPSFAGMVDPSL” to the binding groove of HLA-DRB1*01:01 (affinity: − 7.0 kcal/mol). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)