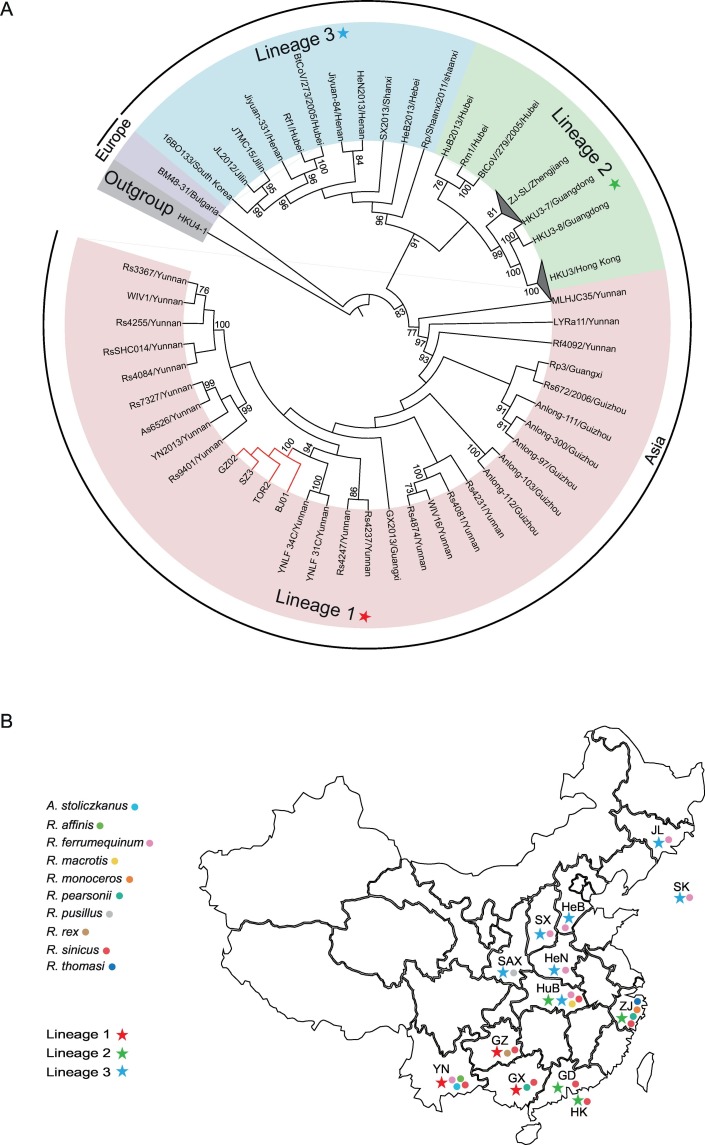
Fig. 1.
Phylogenetic analysis of SARS-CoVs and bat SARSr-CoVs. (A) The phylogenetic tree was constructed using the complete RdRp coding sequences and viewed in iTOL (http://itol.embl.de/). All strains here were named using abbreviations of virus ID and sampling provinces. The strain HKU4-1 (NC_009019) was used as a outgroup of that tree. The taxa for lineage 1, lineage 2 and lineage 3 are highlighted in light red, light green and light blue, respectively. The lineage 1, lineage 2 and lineage 3 are displayed with colored pentagrams. The taxa for the only European strain European BM48–31 (GU190215) is displayed by light purple. The branch of SARS-CoV is marked in red. These strains from Zhejiang were collapsed into a triangle named ZJ-SL/Zhejiang. The viruses from Hong Kong also were collapsed into a triangle named HKU3/Hong Kong. The numbers adjacent to the node represents the bootstrap value of 1000 replicates and only bootstrap values ≥70% are shown. (B) The geographical distribution of bat SARSr-CoVs and their hosts from the lineage 1, lineage 2 and lineage 3 in the A diagram. The South Korea and Chinese provinces from the three lineages are marked in the map using the corresponding colored pentagrams. The bat hosts are also mapped to the corresponding regions in the map. Location abbreviations are as follows: YN, Yunnan; GZ, Guizhou; GX, Guangxi; GD, Guangdong; HK, Hong Kong; SAX, Shaanxi; HuB, Hubei; SX, Shanxi; HeN, Henan; HeB, Hebei; ZJ, Zhejiang; JL, Jilin; SK, South Korea. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)