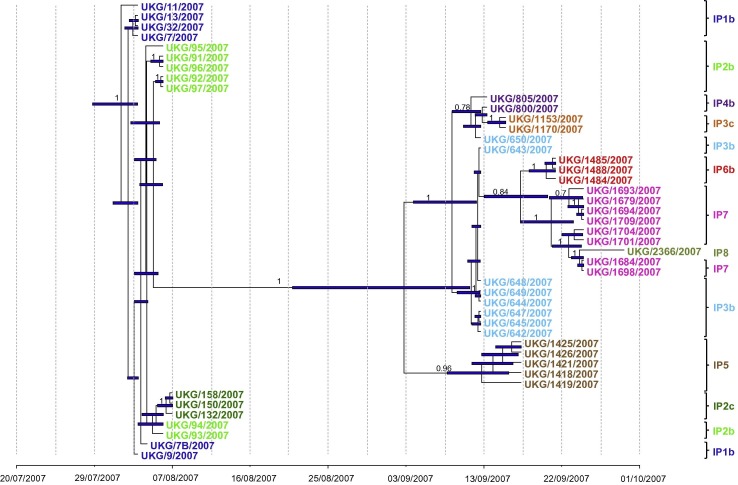
Fig. 1.
Bayesian maximum-clade-credibility time-scaled phylogenetic tree (BEAST) generated using 45 sequenced FMDV full genomes obtained from infected animals during the 2007 outbreak in UK. Sequences from the same holdings are coloured with the same colour as follows: in dark blue, sequences from IP1b; in light and dark green, sequences from IP2b and c, respectively; in brown, sequences from IP5; in light blue and orange, sequences from IP3b and c, respectively; in purple, sequences from IP4b; in red, sequences from IP6; in pink, sequences from IP7; and in green pistachio, sequences from IP8. The analysis was undertaken using the HKY model of base substitution (gamma model of site heterogeneity), exponential relaxed molecular clock, Bayesian skyline plot, sampling 30,000 trees from 30 million generations. Uncertainty for the date of each node (95% highest posterior density – HPD -–intervals) is displayed in bars. Only node labels with posterior over 0.7 are indicated. Overall, a rate of nucleotide substitution of 4.94 × 10−5 (95% HPD: 2.92 × 10−5–7.02 × 10−5) per site per day was estimated. The ancestor is estimated to be on the 01/08/2007 (95% HPD = 29/07/2007–03/08/2007). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)