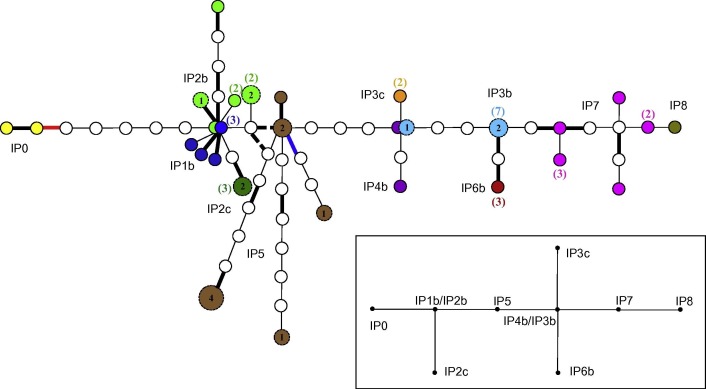
Fig. 2.
Statistical parsimony tree as implemented by TCS using 45 full FMDV genomes obtained from infected animals during the 2007 outbreak in UK. Sequences from the same holdings are coloured with the same colour as per Fig. 1. The sequences in yellow belonged to isolates used at the Pirbright campus during July 2007. When two or more samples within the same premise provided identical sequences, the number of samples represented is shown in brackets in the same colour than the premise to which the sequences belong to. Samples that contained sequence ambiguities are shown as larger labelled circles with the actual number of sites in which ambiguities were present. Lines in bold correspond to non-synonymous nucleotide substitutions. The two intermittent lines represent the two options corresponding to one ambiguity in one site of a virus sequence from IP5. The lines in red correspond to a nucleotide substitution causing an amino acid change (His to Arg) important for heparan sulphate binding (cell culture adaptation). Finally, the lines in blue correspond to nucleotide substitutions causing an amino acid change (Asp to Gly) associated with, but not critical for, heparan sulphate binding. In the right down square a simplified view of the tree (reference tree) was drawn. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)