Highlights
-
•
HCoV−OC43 is involved in healthcare–associated infections.
-
•
HCoV−OC43 genotypes B, E, F and G are identified.
-
•
Asian and European strains of HCoV−OC43 circulate among patients.
Keywords: Human coronavirus, Whole genome sequencing, Hematopoietic cell transplant, Care-associated infections, Phylogenetic analyses
Abstract
Background
While respiratory viral infections are recognized as a frequent cause of illness in hematopoietic stem cell transplantation (HSCT) recipients, HCoV−OC43 infections have rarely been investigated as healthcare-associated infections in this population.
Objectives
In this report, HCoV−OC43 isolates collected from HSCT patients were retrospectively characterized to identify potential clusters of infection that may stand for a hospital transmission.
Study design
Whole-genome and S gene sequences were obtained from nasal swabs using next-generation sequencing and phylogenetic trees were constructed. Similar identity matrix and determination of the most common ancestor were used to compare clusters of patient’s sequences. Amino acids substitutions were analysed.
Results
Genotypes B, E, F and G were identified. Two clusters of patients were defined from chronological data and phylogenetic trees. Analyses of amino acids substitutions of the S protein sequences identified substitutions specific for genotype F strains circulating among European people.
Conclusions
HCoV−OC43 may be implicated in healthcare-associated infections.
1. Background
Although highly pathogenic human coronaviruses (HCoV), like SARS-CoV and MERS-CoV, have emerged recently, the majority of HCoV infections are caused by HCoV-229E, HCoV-NL63, HCoV−OC43 and HCoV-HKU1. These coronaviruses are mainly recognized as causative agents of community-acquired infections. With the exception of SARS and MERS-CoV, there are not many published studies about health-care associated HCoV infections [[1], [2], [3], [4], [5]]. HCoV-229E, HCoV-NL63, HCoV−OC43 and HCoV-HKU1 are usually associated with mild diseases in immunocompetent patients, but can cause severe respiratory tract infections in fragile populations. Indeed, development of molecular detection tests for diagnosis has shown that HCoV are clearly responsible for severe infections in immunocompromised patients, including hematopoietic stem cell transplant recipients (HSCT) [[6], [7], [8]].
HCoV−OC43, HCoV-229E, HCoV-NL63 and HCoV-HKU1 account for 6.7–15.4% of the viruses detected during respiratory infections in HSCT recipients [9]. Currently, HCoV−OC43 is detected around the world, circulating all year long with a slight predominance during winter in temperate countries. The efficient transmission via small droplets and a prolonged shedding by HSCT recipient’s contribute to virus dissemination and highlight the need for a better understanding of hospital-acquired HCoV infections [[10], [11], [12], [13]]. Although an outbreak of HCoV-NL63 respiratory infections in a long-term care facility has recently been reported, a limited amount data are available on the overall genomic characteristics of HCoV−OC43 healthcare-associated infections [14].
The main objective of this work was to identify potential clusters of infection that may stand for a hospital transmission. Next-Generation Sequencing (NGS) was used to generate whole-genome sequences from nasal swabs and phylogenetic analyses were then applied to characterize sequences. The secondary objective was to identify molecular signature of circulating strains.
2. Methods
2.1. Patients
Eight HSCT recipients with HCoV−OC43 detected in nasal swabs by multiplex respiratory viral PCR using Anyplex II RVS 16 detection kit at the University hospital of Lille between 2013 and 2015 were included in the study. Demographic and clinical data were collected. All patients were informed that they were included in a research cohort and agreed that their biological samples could be used for research purposes. Nasal swabs studied here were included into the biological collection of the University Hospital of Lille declared to the French Ministry of High Education and Research (reference number DC-2008-642). According to the French laws, this declaration implies acceptance by an ethic committee. Patients are named as MDSX for “Maladies Du Sang”X.
2.2. Sequencing
Viral genome sequencing was performed as described by Maurier et al. [15]. Briefly, extracted viral RNA was reverse transcribed, PCR amplified using multiple primer pairs and pooled before preparing sequencing libraries using NxSeq AmpFREE LowDNA (Lucigen) library kit and barcoded with Illumina-compatible adaptors. Libraries were paired-end sequenced in 2 × 300 cycles on Illumina MiSeq. Sequencing analysis and genome assembly was performed as described by Maurier et al. [15]
2.3. Genomes annotation
Annotation of the 8 newly sequenced genomes MDS2, MDS4, MDS6, MDS11, MDS12, MDS14, MDS15 and MDS16, was performed by comparison with the HCoV−OC43 isolate from Mexico (Genbank accession KX344031) [16] using BlastN [17]. This isolate served as a reference and allowed us to compute the coordinates of the following functional elements: ORF1a, ORF1b, ns2A, HE, S, M and N genes.
2.4. Phylogenetic analysis
For the whole genome evolutionary analysis of MDS2, MDS4, MDS6, MDS11, MDS12, MDS14, MDS15 and MDS16, we selected 40 published full-length human coronavirus genomes with diverse genotypes and geographical origins, making a total of 48 sequences. We also considered more specifically the phylogeny of S gene and included 12 additional sequences from partially sequenced genomes from Caen hospital (France) and one additional published sequence from China (Table supplementary data), making a total of 61 sequences. For each of the two datasets, three distantly related bovine coronaviruses served as outgroup and multiple-sequence alignment was built using MUSCLE from SeaView 4.6.4 followed by a manual correction step taking into account protein coding sequences for coding regions [18,19]. Based on the multiple sequence alignments, similarity between each pair of nucleotide sequences was computed using the Sequence Manipulation Suite [20]. Phylogenetic trees were then inferred using the maximum likelihood method implemented in PhyML 3.1 with a general time reversible (GTR) nucleotide substitution model, a proportion of invariant sites (+I) and a gamma rate heterogeneity (+Γ) with 1000 bootstrap replicates [21]. The substitution model was chosen by optimizing the AIC score using smart model selection [22]. A total of 30,815 nucleotides positions were included for the whole genome analysis and 4137 nucleotides positions for S gene. The software FigTree 1.4.4 was used to produce figures [23].
2.5. Substitution analysis
Amino acids comparative analyses for ORF1a, ORF1b, ns2A, HE, S, M and N proteins were done on 58 sequences strains (8 MDS + 40 previously included and 10 others) for which complete genomes were available and on 76 sequences for S gene only (8 MDS + 68 others) (Table Supplementary data). For this analysis, we added new sequences compared to the phylogenetic analysis. Indeed, we need to examine each position of the sequences separately, which requires a larger support. By doing this, we reduced the chance to select erroneous mutations. The goal is to ensure that the informative mutations found are characteristic of a particular genotype: they are found only in this genotype and are shared by all sequences known for this genotype. In both cases, the signature amino acid substitutions were determined by selecting a subset of informative sites using DIVEIN [24]. Colorization was performed manually to highlight conserved sites.
2.6. Spatio-temporal and evolutionary dynamics
We followed the methodology used in [25]. Notably, we added Asiatic sequences that were also used in [25] and [26]. We estimated divergence times using BEAST 1.8.4 [12] on both whole genome and S gene [27]. Exponential population size, relaxed clock with coalescent tree (BSP distribution for S and uncorrelated exponential distribution for whole genome) was tested to estimate the time of most recent common ancestor (tMRCA).
All information on the sequences used (identifiers, strains, bibliographical references) is summed up in the supplementary Table S1.
Complete genome sequences were deposited in Genbank under accession numbers MK303619 to MK303625, MK327281
3. Results
3.1. Patients and viral characteristics
HCoV−OC43 isolates were detected by RT-PCR from nasal swabs collected from 8 HSCT recipients. Table 1 shows the characteristics of these patients. The median time to HCoV−OC43 infection after HSCT was 239.5 days (range 13–1052 days). All patients had graft-versus-host diseases for which they received a steroid daily dose of higher than 1 mg/kg, except patient MDS4. All patients died except one, MDS4; however, in all the cases, death was not directly related to HCoV−OC43 infection.
Table 1.
Clinical features of patients with HCoV−OC43 infections.
| Age | Sex | Hematology feature | Transplant numbers | Donor type | Days between transplant and HCoV-infection | date of HCoV detection | Cq value | Respiratory or infectious symptoms | Platelet count | Leukocyte count | Neutrophil count | Lymphocyte count | Monocyte count | steroids | GVH | Co-detection | Outcome | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| y | m/d/y | x10exp9/L | x10exp9 /L | x10exp9 /L | x10exp9 /L | x10exp9/L | ||||||||||||
| MDS-2 | 19 | F | acute myeloid leukemia | 1 | unrelated | 2014 | 3-11-14 | 35 | yes | 196 | 6.45 | 3.6 | 1.3 | 1.4 | y | y | no | death |
| MDS-4 | 30 | M | Multiple myeloma | 2 | auto/unrelated | 13 | 2-15-13 | 34 | yes | 71 | 6.35 | 4.5 | 1 | 0.7 | n | y | no | alive |
| MDS-6 | 64 | F | myeloproliferative syndrome | 1 | unrelated | 359 | 1-22-13 | 37 | yes | 14 | 1.59 | 1 | 0.4 | 0.1 | y | y | no | death |
| MDS-11 | 45 | F | atypical myeloproliferative syndrome | 1 | unrelated | 144 | 1-30-14 | 20 | yes | 21 | 1.49 | 0.3 | 1.1 | 0.1 | y | y | no | death |
| MDS-12 | 65 | F | relaspe of multiple myeloma | 1 | unrelated | 430 | 2-20-14 | 27 | yes | 73 | 1.26 | 0.7 | 0.4 | 0.1 | y | y | Influenza A | death |
| MDS-14 | 33 | F | relapse of lymphoblastic acute leukemia | 2 | unrelated | 335 | 1-8-13 | 25 | yes | 102 | 6.92 | 4.8 | 1.3 | 0.6 | y | y | Influenza A + Bocavirus | death |
| MDS-16 | 33 | M | relaspse of mycosis fungoides | 1 | related | 1052 | 1-16-15 | 27 | yes | 256 | 14.05 | 11.3 | 2.1 | 0.7 | y | y | no | death |
| MDS 15 | 40 | F | acute myeloid leukemia | 1 | unrelated | 103 | 2-11-14 | 24 | yes | 125 | 2.04 | 0.5 | 1.1 | 0.5 | y | n | no | |
| Means | 494.4 | 95.3 | 4.5 | 3.0 | 1.0 | 0.5 |
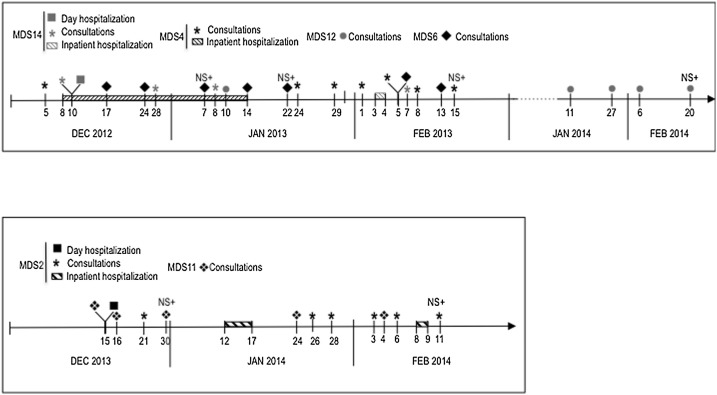
Fig. 1 presents the timeline of events for patients involved in HCoV−OC43 clusters of infections, that why patients MDS15 and MDS16 are not mentioned. Patients were frequently admitted in hospital for consultations in the outpatient clinic or inpatient hospitalizations. They shared the same hospital units and the same healthcare worker teams. At the time of nasal swab collection, acute respiratory symptoms were present for all the patients. Other respiratory pathogens were detected in nasal swabs for only 2 patients (Table 1). No concomitant co-infection with bacteria or fungi was detected. All episodes occurred during winter.
Fig. 1.
Timeline of events. NS+: Nasal swab positive for HCoV−OC43.
3.2. Characteristics of whole genome sequences
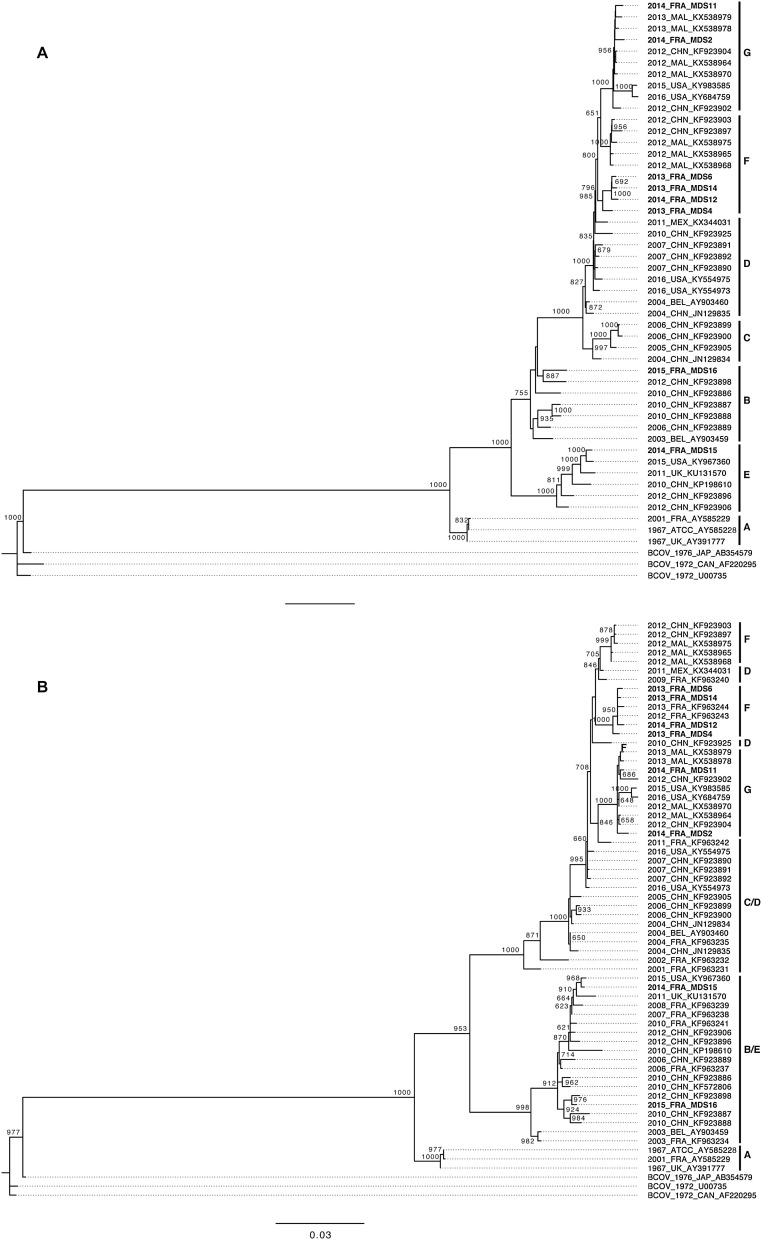
To characterize the overall diversity of HCoV−OC43 circulating among the 8 HSCT patients, 7 full-length genome sequences were obtained from nasal swabs of HSCT patients using NGS method. The sequence obtained from MDS15 patient was incomplete since a part between positions 7473 and 7689 in orf1ab was missing. Considering the small fragment of sequence missing, the partial full-length sequence obtained from this patient was also included in all phylogenetic analyses. To genotype the HCoV−OC43 isolates, phylogenetic trees were reconstructed by the maximum-likelihood method using sequences of full-length genomes and S genes obtained and compared to those retrieved from GenBank (Fig. 2 A/B). The tree obtained from full-length genomes showed that MDS sequences are divided into 4 known genotypes B, F and G (Fig. 2A) [25,26,28]. MDS4, MDS6, MDS14 and MDS12 sequences, belonging to the genotype F, are grouped into the same cluster separated from other sequences of genotype F with high bootstrap value (79.6 %). These sequences were obtained from samples collected in 2013 except for the MDS12 sequence that was obtained from sample collected in 2014. This cluster was named “cluster 2013” (Fig. 2A). Among the cluster 2013, MDS6 and MDS14 sequences are closely related (69.2 % bootstrap value), MDS12 sequence is separated from the MDS 6 and MDS14 sequences (100 % bootstrap value) and MDS4 sequence is separated from the 3 other sequences (98.5 % bootstrap value). On the tree, MDS2 and MDS11 sequences are divided within the sequences of genotype G with too low bootstrap values to separate firmly the sequences of this cluster (Fig. 2A). MDS2 and MDS11 sequences were collected during the year 2014 and will be named cluster 2014. Overall, the tree obtained from S gene sequences shows the same distribution of MDS sequences. Notably, the cluster 2013 appears closely related with previous published sequences collected from French patients in 2012 and 2013. In sum, the HCoV−OC43 sequences obtained from clinical samples in this study belong to genotypes B, E, F and G, and 6 of them appear clustered into two distinct clusters.
Fig. 2.
Maximum-likelihood trees of HCoV−OC43 strains. A, B, C, D, E, F, G genotypes/clades were represented. MDS sequences are in bold. A. Tree obtained from whole-genome sequences. B. Tree obtained from S gene sequences.
The sequences of the cluster 2013 are very conserved at the nucleotide level of the full-length genomes (up to 99 %). As observed on the tree, MDS 6 and 14 sequences are the closest (with 99.93 % of identity), MDS12 sequence is less similar to the MDS6 and MDS14 sequences (99.11 % of identity) and the MDS4 sequence is the most divergent from the other of the cluster 2013 (99 % of identity). However, MDS4 sequence presents a high level of homology with MDS6 and MD14 sequences (99.81 %). Likewise, MDS2 and MDS11 sequences are very similar with 99.86 % of identities. MDS15 presents a range of homologies comprise between 95.4 % and 96.2 % and MDS16 between 98.4 and 99.2 % with the sequences of the clusters 2013 and 2014. These data reinforce the results of the phylogenetic trees and suggest that sequences of each cluster share the same origin.
To reinforce the hypothesis of common origin of clinical isolates, the divergence times of MDS sequences were estimated using BEAST method. The estimated mean evolutionary rate was 4.0 × 10−4 (3.4 × 10−4 – 4.5 × 10−4) substitutions/site/year for the full-length genome and, for the S gene, 5.5 × 10−4 (4.5 × 10-4 – 6.5 × 10−4) substitutions/site/year. Based on the full-length genome and the S gene data, the time of emergence for genotype A was estimated in the 1960s and in the late 1990s to early 2000s for genotypes B to D (Table 2 ). Similarly, emergence of genotypes D to G was estimated in the late 2000s to early 2010s. Based on the evolutionary estimates of the full-length genome and the S gene, the common ancestor of the sequences of the cluster 2013 was dated back to 2010.9 (2009.6–2012.2) and to 2010.9 (2009.8–2011.7), respectively. Furthermore, the common ancestor of the sequences of the cluster 2014 was dated back to 2011.5 (2010.7–2012.5) and to 2011.0 (2010.1–2011.7) for the full-length genome and the S gene, respectively. These estimates obtained from two different databases were consistent and show that the strains of the clusters 2013 emerged from a common ancestor in 2010.9 and those of 2014 emerged from another in 2011 +/-0.5.
Table 2.
Time of the most recent common ancestors (tMRCA) with 95 % highest posterior density (95 % HPD) for HCoV−OC43 genotypes A to G and clusters based on the spike (S) gene and full-length genome.
| tMRCA (95 % HPD) |
||
|---|---|---|
| Genotype | S gene | Full-length genome |
| A | 1963.1 (1956.6–1966.7) | 1967.0 (1966.9–1967.1) |
| B | 1999.3 (1995.1–2002.3) | 2001.6 (2000.3–2002.7) |
| C | 1993.2 (1988.7–1996.5) | 2002.9 (2001.8–2003.7) |
| D | 1998.1 (1994.8–2001.2) | 2003.0 (2002.0–2003.7) |
| E | 2006.5 (2005.7–2007.5) | 2007.2 (2005.9–2008.4) |
| F | 2007.7 (2007.0–2008.3) | 2009.4 (2008.0–2010.5) |
| G | 2010.8 (2010.0–2011.4) | 2010.9 (2010.3–2011.5) |
| Cluster 2013 | 2010.9 (2009.8–2011.7) | 2010.9 (2009.6–2012.2) |
| Cluster 2014 | 2011.0 (2010.1–2011.7) | 2011.5 (2010.7–2012.5) |
| Mean evolutionary rate (nt subst/site/year) | 5.5 × 10−4 (4.5 × 10−4 – 6.5 × 10−4) | 4.0 × 10−4 (3.4 × 10−4 – 4.5 × 10−4) |
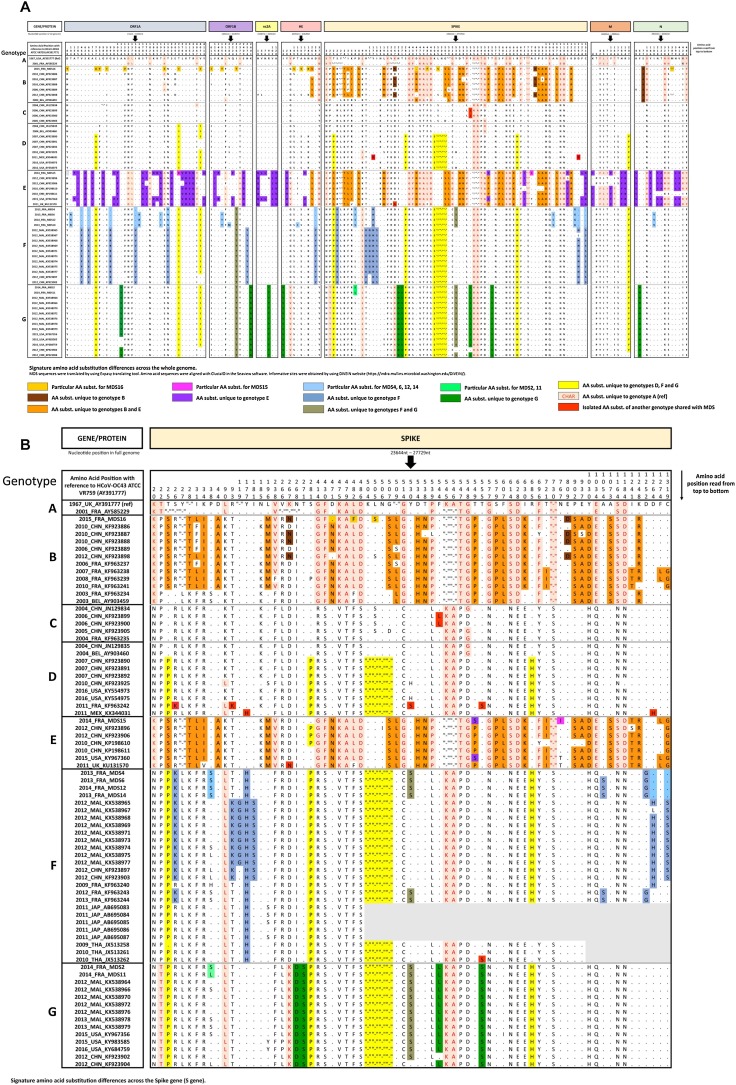
Using the prototype ATCC VR759 as the reference strain for amino acid positions, sequence MDS16 shares 19 amino acid substitutions with other sequences of genotype B and presents 18 specific amino acid substitutions mapped across the whole genome relative to the other sequences of the genotype B (Fig. 3 A). Likewise, sequences of the cluster 2013 show the specific substitution of the genotype F (Y177 H) and 9 additional amino acids substitutions relative to the others of the genotype F [25]. Notably, MDS6, MDS12 and MDS14 sequences show 12 additional substitutions relative to the genotype F, which are absent on the MDS4 sequence. MDS15 sequence, for which only the S region could be analyzed, shares the same amino acid substitutions with the other sequences of the genotype E and shows only 1 specific substitution relative to the other sequences of the same genotype. MDS2 and MDS11 sequences appear similar to the others of genotype G.
Fig. 3.
Signature amino acid substitution differences. A. Amino acid substitution differences across the full-length genome. B. Amino acid substitution differences among the S gene sequences.
3.3. Characteristics of HCoV−OC43 strains
Since S protein sequences of other French HCoV−OC43 isolates have been previously published, phylogenetic analyses show that MDS sequences of the cluster 2013 are clustered to other French sequences on the phylogenetic tree, whereas sequences of the cluster 2014 appear more related to Malaysian, Chinese and American sequences (Fig. 2B) [29]. When signature amino acid substitutions were analyzed, for the genotype B, 3 amino acid substitutions appear on the sequence MDS16: absence of substitution D471 N, substitutions L482 F that is common to other sequences of genotype B, C, F and G and L505S that is also observed on sequences of genotype C. For the genotype E, the amino acid substitution N767I is observed on the sequence MDS15 and had never been described previously. For the genotype F, the amino acid substitution P38S is observed on the MDS4, MDS6, MDS12 and MDS14 sequences which are shared with genotype B and G. The substitutions D1264H and C1319S are absent from the MDS4, MDS6, MDS12 and MDS14 sequences as from the other French sequences K963240, K963243 and K963244. Finally, 2 amino acid substitutions (A1054S and D1252 G) are observed on MDS6, MDS12 and MDS14 sequences and also on previously published sequences K963243 and K963244 (Fig. 3B).
These data show that any amino acid substitution appear specific to sequences of genotype B and G circulating in France. But, sequences circulating among French people belonging to the genotype F had a peculiar molecular signature (absence of substitution D1264H and C1319S, substitution A1054S and D1252 G).
4. Discussion
In this work, we describe retrospectively 8 HCoV−OC43 infections having occurred during the winters 2013 and 2014 at the HSCT Unit of the Lille University hospital. Six patients were divided into 2 clusters, respectively named 2013 and 2014 on the basis of the year of occurrence and genomic characteristics.
HCoV−OC43 cases described in this report could be classified as healthcare-associated infections defined as a group of infections emerging among patients who come from a community with a history of previous exposure to healthcare, but do not fit the nosocomial infection criteria [30]. The 6 patients divided into the two clusters of cases regularly attended the hospital for in- and outpatient cares, as consultations, day and inpatient hospitalizations. So, they were frequently in contact with health care professionals and hospital visitors that may be considered as multiple sources of HCoV infections, during periods of usual circulation of HCoV−OC43 in France [31]. Outside the hospital, they resided in geographically non-related places and no common activity could be identified. So, the hospital admission seems the only link between the patients, even though we were unable to identify any direct contact between the patients. HCoV−OC43 can be transmitted either by an asymptomatic patient with prolonged shedding, by healthcare worker asymptomatic or with an upper respiratory tract infection [10,32]. Phylogenetic analyses showed that whole-genome sequences included in a same cluster were very closely related, had very high levels of similarities and shared same common ancestors. All these elements provide additional evidence that these infections could be classified as healthcare-associated infections. Similarly, to what is observed for influenza viruses, parainfluenza virus type 3 and respiratory syncytial virus, our data show that HCoV−OC43 is involved in healthcare-associated infections [[33], [34], [35]].
Because we were unable to identify the index case of HCoV−OC43 infections of the cluster 2013, the MDS12 patient infection during the winter 2014 with a strain highly related to those circulating the previous year is surprising. However, previous studies have shown that genomes of other high and low pathogenic coronaviruses presented low variation rates during hospital-acquired outbreaks [36,37]. Our data show that sequences of HCoV−OC43 appear also very stable.
Phylogenetic trees based on the whole genome and S gene HCoV−OC43 show that genotypes B, G, E and F currently circulate among French patients, in agreement with other report [29]. Even if HCoV−OC43 was previously detected among patients with haematology malignancy, the involvement of HCoV−OC43 genotypes in this type of infection was rarely investigated [[38], [39], [40]]. Indeed, the analysis of more HCoV−OC43 strains from other healthcare-associated infections will reveal the relative prevalence of each genotype in this type of infections in different localities.
The divergence time of sequences included in the phylogenetic analyses was calculated to accumulate to collect additional information showing the common origin of clinical isolates. Our results are similar to those previously described, validating the tMRCA method used [25,28]. Indeed, our results show that each cluster of sequences has diverged from a distinct ancestor in 2010.9 for the cluster 2013 and in 2011.5 for the cluster 2014.
Due to limited HCoV−OC43 full-length genome published in databases, phylogenetic analyses were conducted on S gene sequences. All results obtained from whole genome and S gene sequences were similar. However, phylogenetic analyses from the numerous published S gene sequences allowed comparison of HCoV−OC43 sequences isolated in Lille with those isolated in different parts of the world. Strains described here represent probably a part of strains currently circulating among European people. However, our data show that both kinds of HCoV−OC43 strains circulate among patients included in the study: those belonging to the genotype G closely related to Asian strains and other belonging to the genotype F more related to other European strains previously described [25,26,28,29,41]. Amino acid substitution analyses showed that some substitutions especially in the S protein could be specific of strains circulating in European population but the number of strains analyzed here and the number of sequences obtained from European isolates of HCoV−OC43 are too small and further studies will be needed to confirm our data.
We are aware that our study presents some limitations. As previously mentioned, we were unable to identify the index case of the HCoV−OC43 infections in the HSCT unit and determine precisely the contact history of the patients. Thus, we cannot exclude that other patients were involved in the HCoV−OC43 healthcare-associated infections described. In the same way, we were unable to detect infection or asymptomatic portage in healthcare workers of the HSCT unit. Some reports have shown that different HCoV−OC43 strains circulated during a same winter, suggesting that there may be multiple channels of HCoV introduction in hospital [25,42,43]. In fact, the HCoV strains of healthcare-associated infections were probably contracted from one of the strains circulating in the community at that time. For all these reasons, cases reported in this study did not correspond to the definition of nosocomial transmission of pathogens and we used the term of coronavirus healthcare-associated infections.
Another weakness of our work concerns the determination of pattern of amino acid substitutions specific of strains circulating among French people. Indeed, our data suggest that some substitutions might be specific to strains circulating in Europe but the analysis of a larger number of strains would be necessary to confirm our hypothesis.
To conclude, we retrospectively described the molecular investigation of HCoV−OC43 infections in a HSCT unit using whole-genome sequencing combined with advanced phylogenetic analyses. These tools allowed us to associate HCoV−OC43 infections with healthcare. Moreover, we suggest that two kinds of HCoV−OC43 strains circulate among the French population, one sharing common ancestors with Asian strains and the other closely related to European lineage.
Funding sources
This work was supported by a Lille Hospital University grant (EPI-CoV) and by CNRS and Lille University (PEPS 2015).
Transparency document
CRediT authorship contribution statement
Delphine Beury: Investigations, Formal analyses. Léa Fléchon: Investigations, Formal analyses. Florence Maurier: Investigations, Formal analyses, Data curation. Ségolène Caboche: Conceptualization, Investigations, Data curation, Writing - original draft. Jean-Stéphane Varré: Conceptualization, Data curation, Writing - original draft. Hélène Touzet: Conceptualization, Data curation, Writing - original draft. Karine Faure: Conceptualization, Resources, Writing - original draft. Jean Dubuisson: Conceptualization, Writing - review & editing, Funding acquisition. David Hot: Conceptualization, Data curation, Writing - original draft. Benoit Guery: Conceptualization, Writing - review & editing, Funding acquisition. Anne Goffard: Conceptualization, Resources, Writing - original draft, Funding acquisition, Project administration.
Declaration of Competing Interest
The manuscript had not been published or presented elsewhere, and the authors have no conflict of interest to declare.
Acknowledgement
The authors thank J. Ogiez for the management of the biological collection.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2019.104206.
Contributor Information
Delphine Beury, Email: delphine.beury@pasteur-lille.fr.
Léa Fléchon, Email: lea.flechon.etu@univ-lille.fr.
Florence Maurier, Email: florence.maurier@pasteur-lille.fr.
Ségolène Caboche, Email: segolene.caboche@pasteur-lille.fr.
Jean-Stéphane Varré, Email: jean-stephane.varre@lifl.fr.
Hélène Touzet, Email: helene.touzet@lifl.fr.
Karine Faure, Email: karine.faure@univ-lille.fr.
Jean Dubuisson, Email: jean.dubuisson@ibl.cnrs.fr.
David Hot, Email: david.hot@pasteur-lille.fr.
Benoit Guery, Email: Benoit.Guery@unil.ch.
Anne Goffard, Email: anne.goffard@univ-lille.fr.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Gagneur A., Sizun J., Vallet S., Legr M.C., Picard B., Talbot P.J. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J. Hosp. Infect. 2002;51:59–64. doi: 10.1053/jhin.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Gagneur A., Vallet S., Talbot P.J., Legrand-Quillien M.-C., Picard B., Payan C., Sizun J. Outbreaks of human coronavirus in a pediatric and neonatal intensive care unit. Eur. J. Pediatr. 2008;167:1427–1434. doi: 10.1007/s00431-008-0687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alhamlan F.S., Majumder M.S., Brownstein J.S., Hawkins J., Al-Abdely H.M., Alzahrani A., Obaid D.A., Al-Ahdal M.N., BinSaeed A. Case characteristics among Middle East respiratory syndrome coronavirus outbreak and non-outbreak cases in Saudi Arabia from 2012 to 2015. BMJ Open. 2017;7 doi: 10.1136/bmjopen-2016-011865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majumder M.S., Brownstein J.S., Finkelstein S.N., Larson R.C., Bourouiba L. Nosocomial amplification of MERS-coronavirus in South Korea, 2015. Trans. R. Soc. Trop. Med. Hyg. 2017;111:261–269. doi: 10.1093/trstmh/trx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbino J., Crespo S., Aubert J.-D., Rochat T., Ninet B., Deffernez C., Wunderli W., Pache J.-C., Soccal P.M., Kaiser L. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (Non-SARS)-related human coronavirus infection. Clin. Infect. Dis. 2006;43:1009–1015. doi: 10.1086/507898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakki M., Rattray R.M., Press R.D. The clinical impact of coronavirus infection in patients with hematologic malignancies and hematopoietic stem cell transplant recipients. J. Clin. Virol. 2015;68:1–5. doi: 10.1016/j.jcv.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piñana J.L., Madrid S., Pérez A., Hernández-Boluda J.C., Giménez E., Terol M.J., Calabuig M., Navarro D., Solano C. Epidemiologic and clinical characteristics of coronavirus and bocavirus respiratory infections after allogeneic stem cell transplantation: a prospective single-center study. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2018;24:563–570. doi: 10.1016/j.bbmt.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milano F., Campbell A.P., Guthrie K.A., Kuypers J., Englund J.A., Corey L., Boeckh M. Human rhinovirus and coronavirus detection among allogeneic hematopoietic stem cell transplantation recipients. Blood. 2010;115:2088–2094. doi: 10.1182/blood-2009-09-244152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogimi C., Greninger A.L., Waghmare A.A., Kuypers J.M., Shean R.C., Xie H., Leisenring W.M., Stevens-Ayers T.L., Jerome K.R., Englund J.A., Boeckh M. Prolonged shedding of human coronavirus in hematopoietic cell transplant recipients: risk factors and viral genome evolution. J. Infect. Dis. 2017;216:203–209. doi: 10.1093/infdis/jix264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerna G., Campanini G., Rovida F., Percivalle E., Sarasini A., Marchi A., Baldanti F. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J. Med. Virol. 2006;78:938–949. doi: 10.1002/jmv.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackay I.M., Arden K.E., Speicher D.J., O’Neil N.T., McErlean P.K., Greer R.M., Nissen M.D., Sloots T.P. Co-circulation of four human coronaviruses (HCoVs) in Queensland children with acute respiratory tract illnesses in 2004. Viruses. 2012;4:637–653. doi: 10.3390/v4040637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva C.S., Mullis L.B., Pereira O., Saif L.J., Vlasova A., Zhang X., Owens R.J., Paulson D., Taylor D., Haynes L.M., Azevedo M.P. Human Respiratory Coronaviruses Detected In Patients with Influenza-Like Illness in Arkansas, USA. Virol. Mycol. Infect. Dis. 2014;2014 doi: 10.4172/2161-0517.S2-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. Hand, EB. Rose, A. Salinas, X. Lu, SK Sakthivel, E. Schneider, JT. Watson. Severe Respiratory Illness Outbreak Associated with Human Coronavirus NL63 in a Long-Term Care Facility - Volume 24, Number 10—October 2018 - Emerging Infectious Diseases journal - CDC, (n.d.). doi:10.3201/eid2410.180862. [DOI] [PMC free article] [PubMed]
- 15.Maurier F., Beury D., Fléchon L., Varré J.-S., Touzet H., Goffard A., Hot D., Caboche S. A complete protocol for whole-genome sequencing of virus from clinical samples: application to coronavirus OC43. Virology. 2019;531:141–148. doi: 10.1016/j.virol.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taboada B.T., Isa P., Espinoza M.A., Aponte F.E., Arias-Ortiz M.A., Monge-Martínez J., Rodríguez-Vázquez R., Díaz-Hernández F., Zárate-Vidal F., Wong-Chew R.M., Firo-Reyes V., Del Río-Almendárez C.N., Gaitán-Meza J., Villaseñor-Sierra A., Martínez-Aguilar G., García-Borjas M., Noyola D.E., Pérez-Gónzalez L.F., López S., Santos-Preciado J.I., Arias C.F. Complete genome sequence of human coronavirus OC43 isolated from Mexico. Genome Announc. 2016;4 doi: 10.1128/genomeA.01256-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 20.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28 doi: 10.2144/00286ir01. 1102, 1104. [DOI] [PubMed] [Google Scholar]
- 21.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 22.Lefort V., Longueville J.-E., Gascuel O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FigTree, (n.d.). http://tree.bio.ed.ac.uk/software/figtree/ (accessed January 28, 2019).
- 24.Deng W., Maust B.S., Nickle D.C., Learn G.H., Liu Y., Heath L., Kosakovsky Pond S.L., Mullins J.I. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. BioTechniques. 2010;48:405–408. doi: 10.2144/000113370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oong X.Y., Ng K.T., Takebe Y., Ng L.J., Chan K.G., Chook J.B., Kamarulzaman A., Tee K.K. Identification and evolutionary dynamics of two novel human coronavirus OC43 genotypes associated with acute respiratory infections: phylogenetic, spatiotemporal and transmission network analyses. Emerg. Microbes Infect. 2017;6:e3. doi: 10.1038/emi.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Li J., Xiao Y., Zhang J., Wang Y., Chen L., Paranhos-Baccalà G., Ren L., Wang J. Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination. J. Infect. 2015;70:641–650. doi: 10.1016/j.jinf.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau S.K.P., Lee P., Tsang A.K.L., Yip C.C.Y., Tse H., Lee R.A., So L.-Y., Lau Y.-L., Chan K.-H., Woo P.C.Y., Yuen K.-Y. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kin N., Miszczak F., Lin W., Gouilh M.A., Vabret A., Consortium E. Genomic analysis of 15 human coronaviruses OC43 (HCoV-OC43s) circulating in France from 2001 to 2013 reveals a high intra-specific diversity with new recombinant genotypes. Viruses. 2015;7:2358–2377. doi: 10.3390/v7052358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardoso T., Almeida M., Friedman N.D., Aragão I., Costa-Pereira A., Sarmento A.E., Azevedo L. Classification of healthcare-associated infection: a systematic review 10 years after the first proposal. BMC Med. 2014;12:40. doi: 10.1186/1741-7015-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassoun A., Huff M.D., Weisman D., Chahal K., Asis E., Stalons D., Grigorenko E., Green J., Malone L.L., Clemmons S., Lu S. Seasonal variation of respiratory pathogen colonization in asymptomatic health care professionals: a single-center, cross-sectional, 2-season observational study. Am. J. Infect. Control. 2015;43:865–870. doi: 10.1016/j.ajic.2015.04.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esbenshade J.C., Edwards K.M., Esbenshade A.J., Rodriguez V.E., Talbot H.K., Joseph M.F., Nwosu S.K., Chappell J.D., Gern J.E., Williams J.V., Talbot T.R. Respiratory virus shedding in a cohort of on-duty healthcare workers undergoing prospective surveillance. Infect. Control Hosp. Epidemiol. 2013;34:373–378. doi: 10.1086/669857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J.H., Choi E.H., Kang H.J., Park K.D., Park S.S., Shin H.Y., Lee H.J., Ahn H.S. Respiratory viral infections after hematopoietic stem cell transplantation in children. J. Korean Med. Sci. 2013;28:36–41. doi: 10.3346/jkms.2013.28.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weedon K.M., Rupp A.H., Heffron A.C., Kelly S.F., Zheng X., Shulman S.T., Gutman P., Wang D., Zhou Y., Noskin G.A., Anderson E.J. The impact of infection control upon hospital-acquired influenza and respiratory syncytial virus. Scand. J. Infect. Dis. 2013;45:297–303. doi: 10.3109/00365548.2012.726738. [DOI] [PubMed] [Google Scholar]
- 35.Loubet P., Voiriot G., Houhou-Fidouh N., Neuville M., Bouadma L., Lescure F.-X., Descamps D., Timsit J.-F., Yazdanpanah Y., Visseaux B. Impact of respiratory viruses in hospital-acquired pneumonia in the intensive care unit: a single-center retrospective study. J. Clin. Virol. 2017;91:52–57. doi: 10.1016/j.jcv.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hays J.P., Myint S.H. PCR sequencing of the spike genes of geographically and chronologically distinct human coronaviruses 229E. J. Virol. Methods. 1998;75:179–193. doi: 10.1016/S0166-0934(98)00116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong S., Lingappa J.R., Chen Q., Shu B., LaMonte A.C., Cook B.T., Birge C., Wang Chern S., Liu X., Galloway R., Le Quynh M., Wai F.N., Yang J.-Y., Butany J., Comer J.A., Monroe S.S., Beard S.R., Ksiazek T.G., Erdman D., Rota P.A., Pallansch M.A., Anderson L.J. Direct sequencing of SARS-Coronavirus S and N genes from clinical specimens shows limited variation. J. Infect. Dis. 2004;190:1127–1131. doi: 10.1086/422849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikulska M., Del Bono V., Gandolfo N., Dini S., Dominietto A., Di Grazia C., Bregante S., Varaldo R., Orsi A., Ansaldi F., Bacigalupo A., Viscoli C. Epidemiology of viral respiratory tract infections in an outpatient haematology facility. Ann. Hematol. 2014;93:669–676. doi: 10.1007/s00277-013-1912-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutspardol S., Essa M., Richardson S., Schechter T., Ali M., Krueger J., Fujii H., Egeler R.M., Gassas A. Significant transplantation-related mortality from respiratory virus infections within the first one hundred days in children after hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2015;21:1802–1807. doi: 10.1016/j.bbmt.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogimi C., Waghmare A.A., Kuypers J.M., Xie H., Yeung C.C., Leisenring W.M., Seo S., Choi S.-M., Jerome K.R., Englund J.A., Boeckh M. Clinical significance of human coronavirus in Bronchoalveolar Lavage samples from hematopoietic cell transplant recipients and patients with hematologic malignancies. Clin. Infect. Dis. 2017;64:1532–1539. doi: 10.1093/cid/cix160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kin N., Miszczak F., Diancourt L., Caro V., Moutou F., Vabret A., Ar Gouilh M. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect. Genet. Evol. 2016;40:186–191. doi: 10.1016/j.meegid.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y., Li C., Chen L., Xu B., Zhou Y., Cao L., Shang Y., Fu Z., Chen A., Deng L., Bao Y., Sun Y., Ning L., Liu C., Yin J., Xie Z., Shen K. A novel human coronavirus OC43 genotype detected in mainland China. Emerg. Microbes Infect. 2018;7 doi: 10.1038/s41426-018-0171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S.-F., Tuo J.-L., Huang X.-B., Zhu X., Zhang D.-M., Zhou K., Yuan L., Luo H.-J., Zheng B.-J., Yuen K.-Y., Li M.-F., Cao K.-Y., Xu L. Epidemiology characteristics of human coronaviruses in patients with respiratory infection symptoms and phylogenetic analysis of HCoV-OC43 during 2010-2015 in Guangzhou. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.