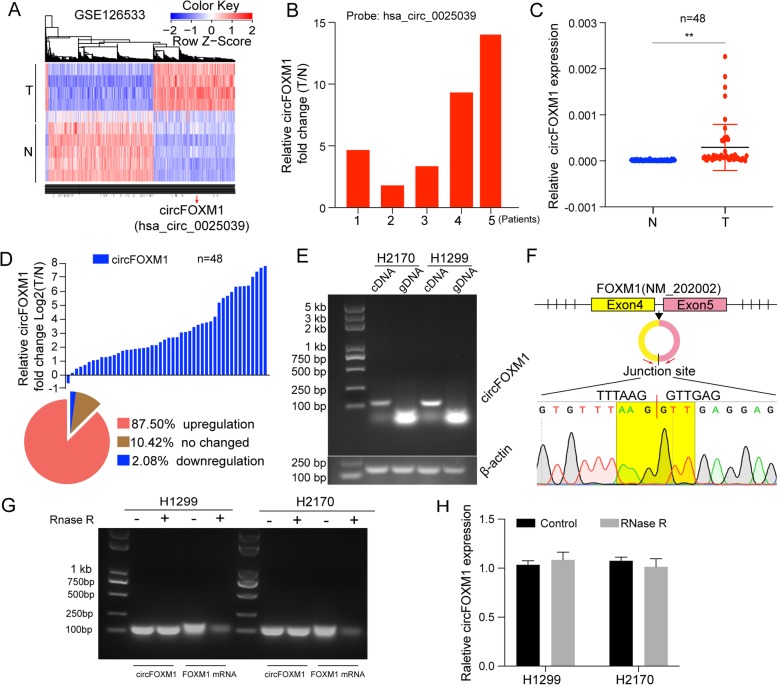
Fig. 1.
Identification and characteristic of circFOXM1 in NSCLC. a circFOXM1 was one of the most upregulated circRNAs based on the analysis of the5 paired samples of NSCLC tissues in GSE126533. T, tumor tissue; N, nontumor tissue. b Relative fold change (T/N) of circFOXM1 in GSE126533. c Expression of circFOXM1 was determined in 48 paired samples of NSCLC tissues. β-actin was used as control. d Histogram and pie chart of the proportions of NSCLC samples in which circFOXM1 expression was upregulated (42/48, 87.50%, red), downregulated (1/48, 2.08%, blue), or no change (5/48, 10.42%, brown). Log2 (T/N) expression value > 1 as higher expression, which < − 1 as lower expression, and between − 1 and 1 as no significant change. e PCR analysis for circFOXM1 in cDNA and genomic DNA (gDNA). f A schematic view of circFOXM1 genomic loci. circFOXM1 is produced at the FOXM1 gene (NM_202002) locus containing exons 4–5. The back-splice junction of circFOXM1 was identified by Sanger sequencing. g RT-PCR analysis for circFOXM1 and FOXM1 mRNA after treatment with RNase R in NSCLC cells. h qRT-PCR analysis for circFOXM1 after treatment with RNase R in NSCLC cells. **P < 0.01