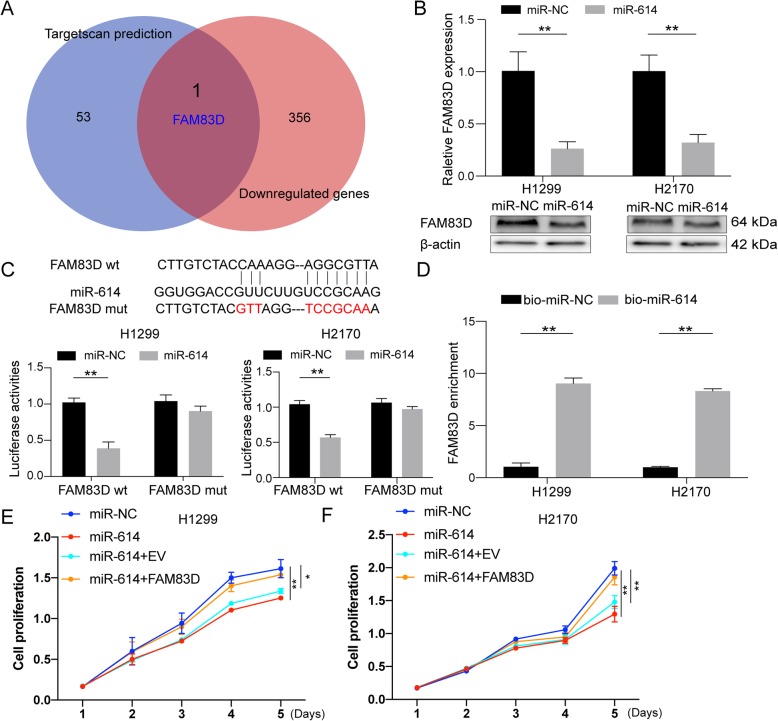
Fig. 5.
miR-614 inhibited cell proliferation via FAM83D. a Venn diagram showed the intersection between miR-614 potential targets predicted by Targetscan (blue circle, 8mer sites) and downregulated genes (red circle, |fold change| > 2, FDR < 0.1) after silencing circFOXM1 in H1299 cells. FAM83D is the only gene in the overlapping of the two datasets. b mRNA and protein levels of FAM83D after transfection of miR-614 or miR-NC in H1299 and H2170 cells. c Upper panel: a schematic illustration showed the putative binding sites of the miRNAs associated with 3’UTR of FAM83D. Lower panel: luciferase activity of 3′ UTR of Luc-FAM83D in H1299 and H2170 cells transfected with miR-614 mimics or miR-NC. d RNA pull-down assay was used for the detection of FAM83D with either miR-614 or miR-NC probe in H1299 and H2170 cells. e-f Cell proliferation analysis of H1299 and H2170 cells transfected with miR-NC, miR-614, miR-614 + empty vector (EV), or miR-614 + FAM83D by CCK-8 assay. *P < 0.05; **P < 0.01