Fig. 1 |. Transcription-associated RNA surveillance.
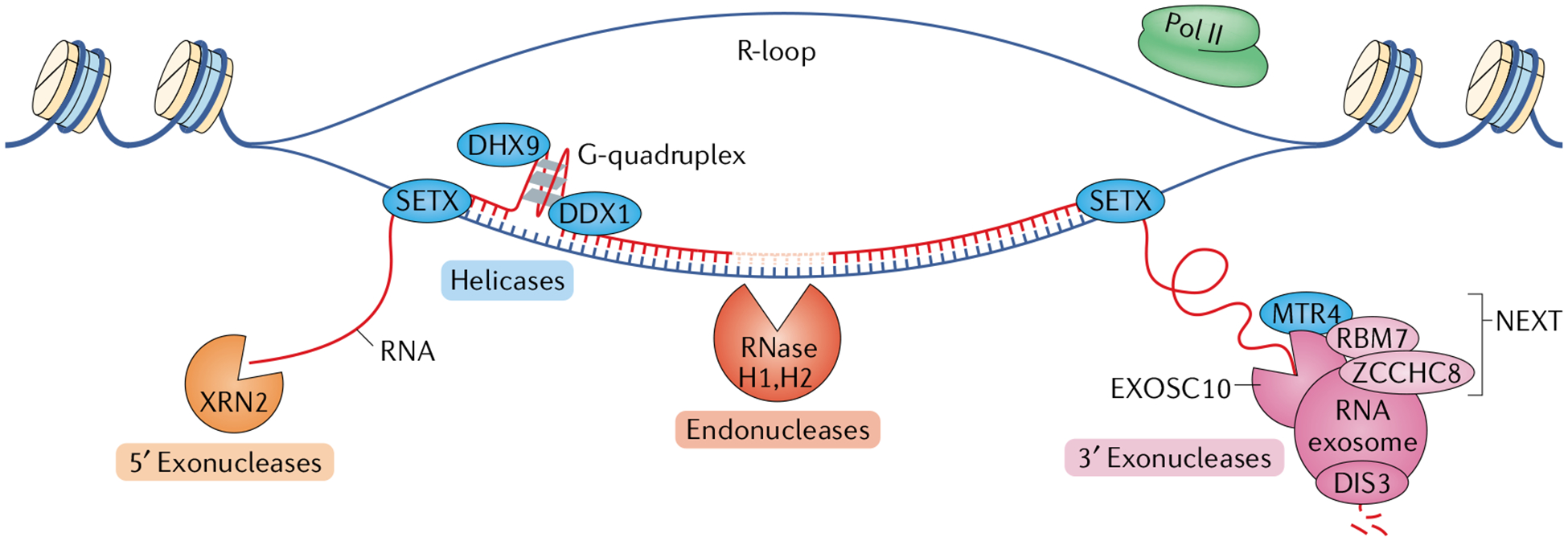
Several components of RNA surveillance are associated with the transcription of both coding and non-coding RNAs; these include factors for RNA degradation and for RNA–DNA and RNA–RNA unwinding that facilitate said degradation (not shown). The three major degradation machineries are 5′ exonucleases, endonucleases and 3′ exonucleases. Depicted here are the components that are most important for transcriptional surveillance. The proteins 5′–3′ exoribonuclease 1 (XRN1) and XRN2 are the primary 5′ exoribonucleases. XRN2 is located in the nucleus, making it most relevant for transcription-associated 5′ exonucleolytic RNA degradation. XRN2 and the helicase senataxin (SETX) function together in mediating transcription termination (not shown). The ribonuclease H (RNase H) enzymes are the primary endoribonucleases in mammalian cells. RNase H1 is located both in the mitochondria and in the nucleus, whereas RNase H2, which is the main source of transcription-associated endonuclease activity, is located only in the nucleus. The 3′ exoribonuclease most tightly associated with transcription of both coding and non-coding RNA is the RNA exosome complex. Its main catalytic subunit is DIS3, though exosome component 10 (EXOSC10) also has catalytic activity. The RNA exosome often associates with the nuclear exosome-targeting (NEXT) complex comprising RNA-binding protein 7 (RBM7), the scaffolding protein zinc finger CCHC domain-containing protein 8 (ZCCHC8) and the RNA helicase MTR4, which is required for efficient RNA exosome activity. In some cases, such as in the process of class switch recombination in B lymphocytes, SETX joins MTR4 in unwinding RNA–DNA and RNA–RNA substrates to promote RNA exosome activity. Several other RNA helicases function in transcription-associated RNA surveillance, including the DEAD-box (DDX) and DEAH-box (DHX) helicases, which help prevent DNA damage. For example, DDX1 and DHX9 can each unwind G-quadruplex structures to facilitate R-loop resolution. Pol II, RNA polymerase II.
