Fig. 2 |. RNA surveillance between deleterious and programmed DNA mutagenesis.
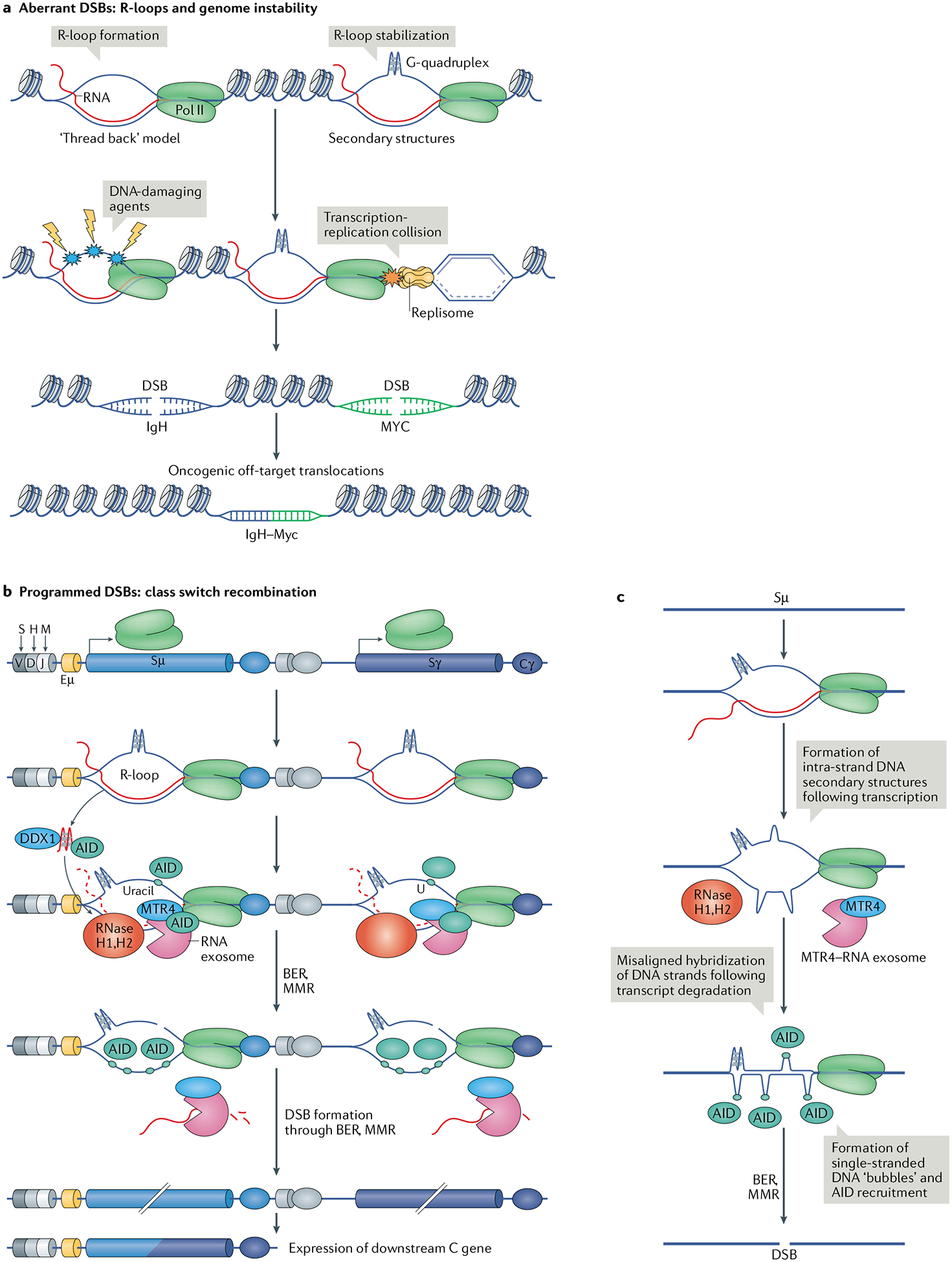
a | The current model of R-loop formation is ‘threading back’ of the nascent RNA into the DNA. R-loops are stabilized by the formation of secondary DNA structures, such as G-quadruplexes, on the exposed single-stranded DNA (ssDNA). R-loop formation and stabilization is a threat to DNA integrity because the ssDNA is vulnerable to DNA-damaging agents. R-loops and the associated RNA polymerase II (Pol II) can also pose a threat during DNA replication in the form of collisions between Pol II and the replisome. Consequently, R-loop persistence increases the threat of formation of aberrant DNA double-stranded breaks (DSBs), which can cause potentially oncogenic DNA translocations such as the common oncogenic translocation between the immunoglobulin heavy chain locus (IgH) and Myc. b | During class switch recombination of the immunoglobulin locus, transcription of non-coding RNA in switch sequences upstream of constant (C) genes leads to the formation of R-loops. The RNA exosome complex and other enzymes are recruited to these R-loops along with activation-induced cytidine deaminase (AID). R-loop formation allows AID to deaminate cytidines on the non-template strand. AID access to the template strand is enabled by the RNA exosome and ribonuclease (RNase) H1 and RNase H2, which degrade the nascent RNA strand of the RNA–DNA hybrid. Owing to the ability of the non-template DNA strand to form stable secondary DNA structures, the two DNA strands are slow to reanneal; this increases the availability of ssDNA substrates for AID and mutagenesis. G-quadruplex-forming, nascent non-coding RNAs at switch sequences are bound by AID, and this binding and RNA degradation by the RNA exosome are enhanced by the unwinding of the RNA by the helicase DEAD-box 1 (DDX1). Processing of AID-associated mutations in the switch sequence by the base excision repair (BER) or mismatch repair (MMR) pathway leads to DSB formation, ultimately resulting in recombination and expression of the downstream constant gene. c | Another possible mechanism of AID recruitment and DSB formation involves transcription in the Sμ segment, which is rich in dG–dC sequences, by Pol II and R-loop formation. The non-transcribed DNA strand, exposed as ssDNA, forms a G-quadruplex that ‘crunches’ the DNA strand and promotes misalignment of the two DNA strands. Upon degradation of the non-coding RNA transcript by the RNA exosome and RNase H enzymes, aberrant dG–dC pairing results in the formation of ‘bubbles’ of ssDNA, which act as AID substrates, thereby resulting in the formation of a DSB.
