Fig. 3 |. Functions of RNA surveillance in DNA damage repair and chromatin biology.
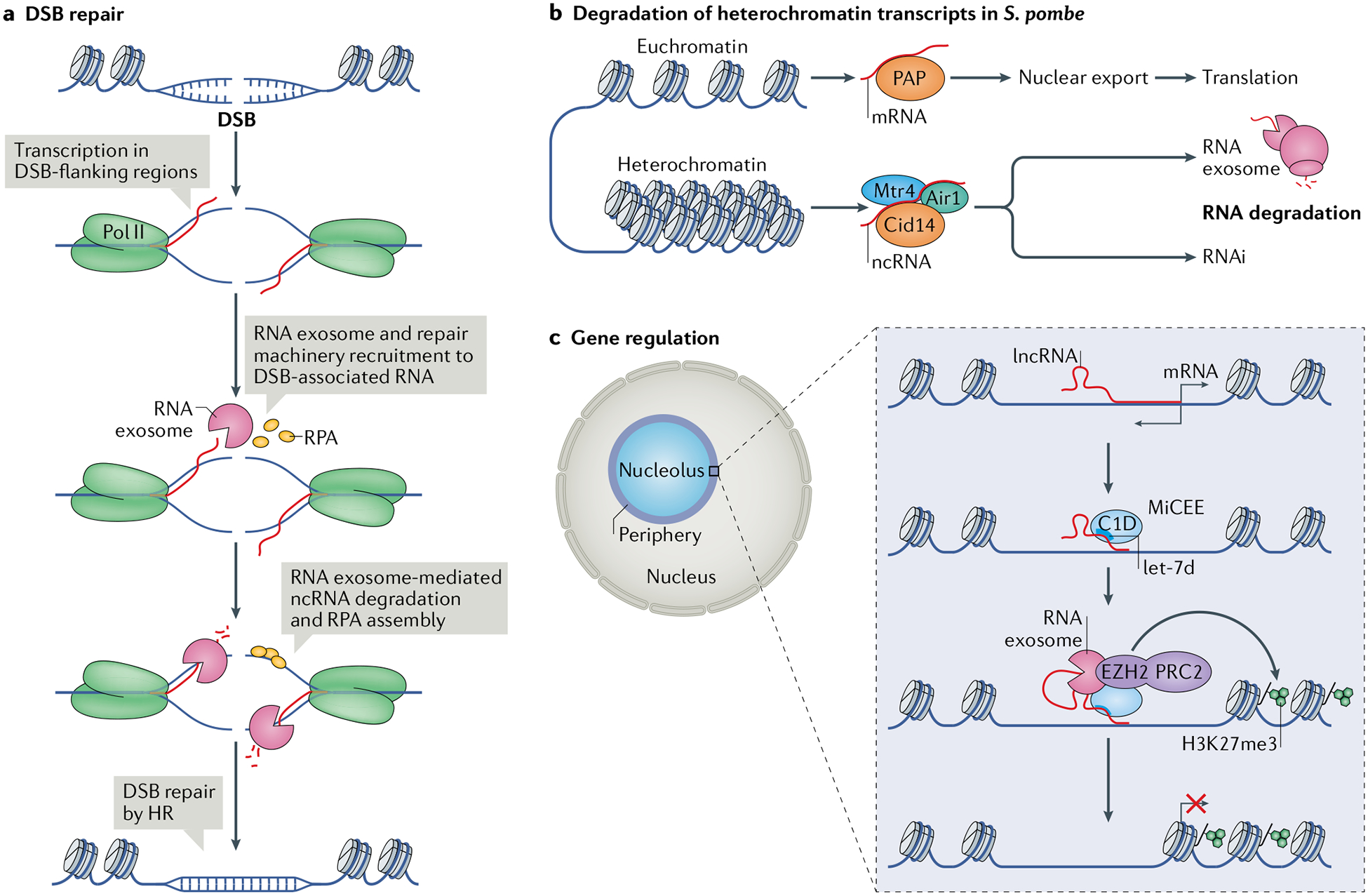
a | RNA surveillance factors such as the RNA exosome can function in DNA double-stranded break (DSB) repair97. Upon DSB formation, the DSB-flanking regions are transcribed and the RNA exosome is recruited to the DSB-associated nascent transcripts along with DNA repair factors such as replication protein A (RPA), which binds to single-stranded DNA (ssDNA). Degradation of the non-coding RNA (ncRNA) by the RNA exosome is coupled with ssDNA binding by RPA, which recruits the homologous recombination (HR) repair machinery and leads to repair of the DSB. b | In the yeast Schizosaccharomyces pombe, the RNA exosome has a role in degrading heterochromatin-derived transcripts in parallel to RNA interference (RNAi)-mediated degradation102. Euchromatin-derived mRNAs undergo processing by poly(A) polymerase (PAP), are exported from the nucleus and undergo translation in the cytoplasm. By contrast, heterochromatin transcripts are processed by the non-canonical poly(A) polymerase Cid14 in association with Mtr4 and Air1. The polyadenylated heterochromatin-derived transcripts are then degraded either by the RNA exosome, the RNAi pathway or both. c | Bidirectional transcription from gene promoters produces, in addition to mRNAs, long non-coding RNAs (lncRNAs) that can be bound by the microRNA let-7d–C1D–EXOSC10–EZH2 (MiCEE) complex. EZH2 is the catalytic component of Polycomb repressive complex (PRC2), which catalyses histone H3 Lys27 trimethylation (H3K27me3) and transcriptional silencing103. Pol II, RNA polymerase II.
