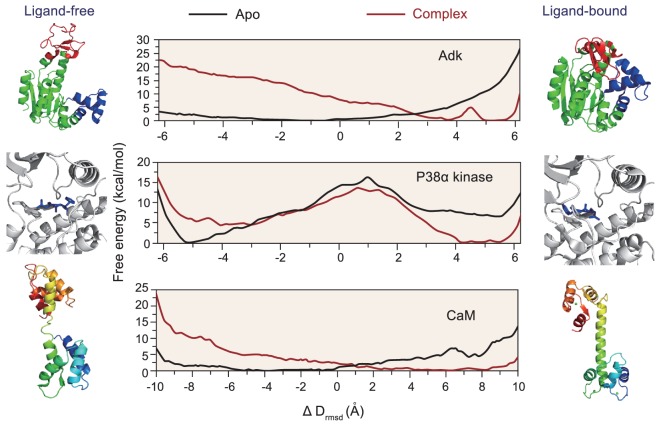
Figure 5.
Schematic representations of NUMD application on three well-known protein systems (adenylate kinase (AdK), calmodulin (CaM), and p38 kinase). The ‘apo’ represents the ligand-free conformation, ‘complex’ means ligand-bound state. The order parameterΔDrmsd is defined as the difference in RMSD values of each structure from the reference starting and final states [80].