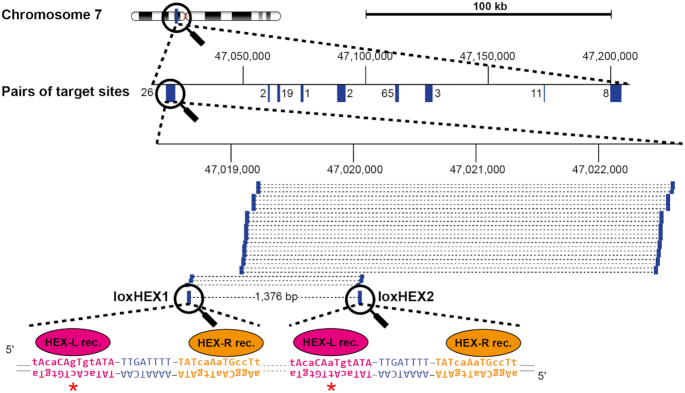
Figure 2.
Nomination of target sites for heterospecific Cre-like recombinases. Bioinformatic analysis of a randomly chosen 200 kb region on chromosome 7 to nominate target sites for dDRiGD. Magnification symbols indicate a zoom-in for the regions of interest. 137 potential target sites cluster in nine different regions, depicted in blue. A zoom into one of these clusters is shown that harbors the selected target sites loxHEX1 and loxHEX2. All pairs of potential target sites located in the cluster are represented as blue bars connected with dashed lines. The selected pair is emphasized by thicker bars, target site names and the distance in base pairs (bp) between them are shown. Numbers above indicate genomic coordinates. The actual DNA sequence of loxHEX1 and loxHEX2 is presented with individual half-sites shown in magenta and orange, with the spacer sequence highlighted in blue. Asymmetric positions are shown in lower case and the red asterisk indicates the single nucleotide difference of the loxHEX1 site compared to the loxHEX2 site. Recombinase monomers (HEX-L rec and HEX-R rec) binding to the individual half-sites are illustrated in the corresponding colors.