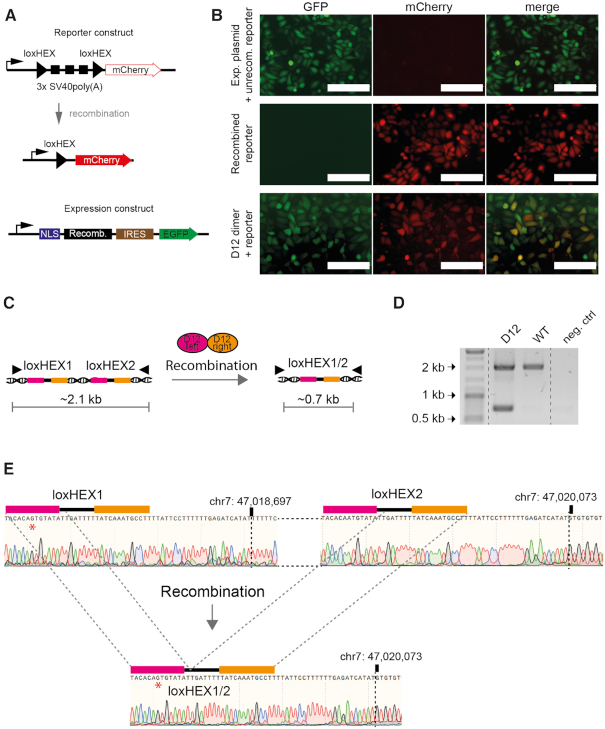
Figure 5.
Recombinase heterodimers are active in mammalian cells. (A) Graphic presentation of the mammalian reporter and expression plasmids. Upon recombination of the reporter construct the 3xSV40poly(A) will be excised allowing for the expression of mCherry (red). Important elements in the reporter and recombinase expression vectors are indicated. Black triangles represent loxHEX target-sites. NLS, nuclear localization signal; Recomb., recombinase coding sequence; IRES, internal ribosome entry site. (B) Fluorescence microscopy images of HeLa cells transfected with the empty expression plasmid and nonrecombined reporter plasmid (top panel), the recombined reporter control (middle panel) or co-transfection of the reporter with the D12 recombinase expression plasmids (lower panel) are shown. Scale bars represent 200 μm. (C) Graphical scheme of the recombination event on genomic DNA. Filled triangles in the illustration represent primer binding sites. After PCR on genomic DNA the nonrecombined locus will be amplified as a ∼2.1 kb fragment whereas the recombined locus will result in a ∼0.7 kb fragment. (D) The D12 heterodimer recombines the endogenous loxHEX locus in human cells. PCR analysis on genomic DNA isolated from cells transfected with D12 and nontransfected control cells are shown. Neg. ctrl. = water control. (E) Sanger-sequencing reads of the WT and recombined PCR fragment on the genomic DNA obtained from WT HeLa cells or transfected HeLa cells with the D12 heterodimer are shown. A repeated region of 23 bp after loxHEX1 and loxHEX2 is indicated. The exact positions on chromosome 7 where the WT differs from the recombined locus is marked with a dotted line. The red asterisk indicates a single nucleotide difference in the loxHEX1 site compared to the loxHEX2 site. A deletion of 1376 bp is observed and demonstrates the precise excision between the two loxHEX target sites.