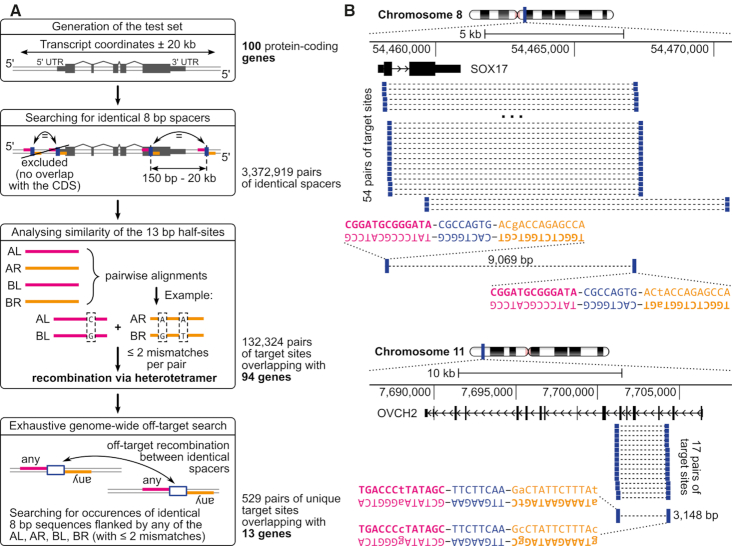
Figure 6.
Evaluation of 100 protein-coding genes in respect to a possibility of performing gene knockouts via dDRiGD. (A) The workflow starts with extracting genomic sequences covering transcript coordinates extended by 20 kb in each direction. In the next step, each sequence is scanned for repeats of identical 8 bp sequences (blue bars) and their 13 bp-flanking sequences (in orange and magenta, AL, AR, BL, BR). Pairs that do not flank coding sequences are discarded. By performing pairwise sequence similarity analysis, half-site sequences are checked to ensure that the target sites can be recombined with two SSRs (no more than two mismatches allowed). In the final step, off-target analyses are performed. Count statistics, on the right of the workflow, indicate abundance of target site pairs generated at each step and counts of genes covered by the target sites. (B) Two example protein coding genes amenable for dDRiGD with their nominated target sites are presented. Each example shows a zoom-in on a chromosomal location, with numbers indicating genomic coordinates, accompanied by a scale bar. Black rectangles connected by arrowed lines represent exons and introns, with arrows indicating direction of transcription. All pairs of potential target sites overlapping with coding sequences (thicker sections of the rectangles) are shown as blue bars connected with dashed lines. For each example, one pair is shown in detail with sequences of target sites and the distance in base pairs (bp) between them. Spacer sequences are highlighted in blue, and half-sites to be bound by the same recombinase are shown in either magenta or orange.