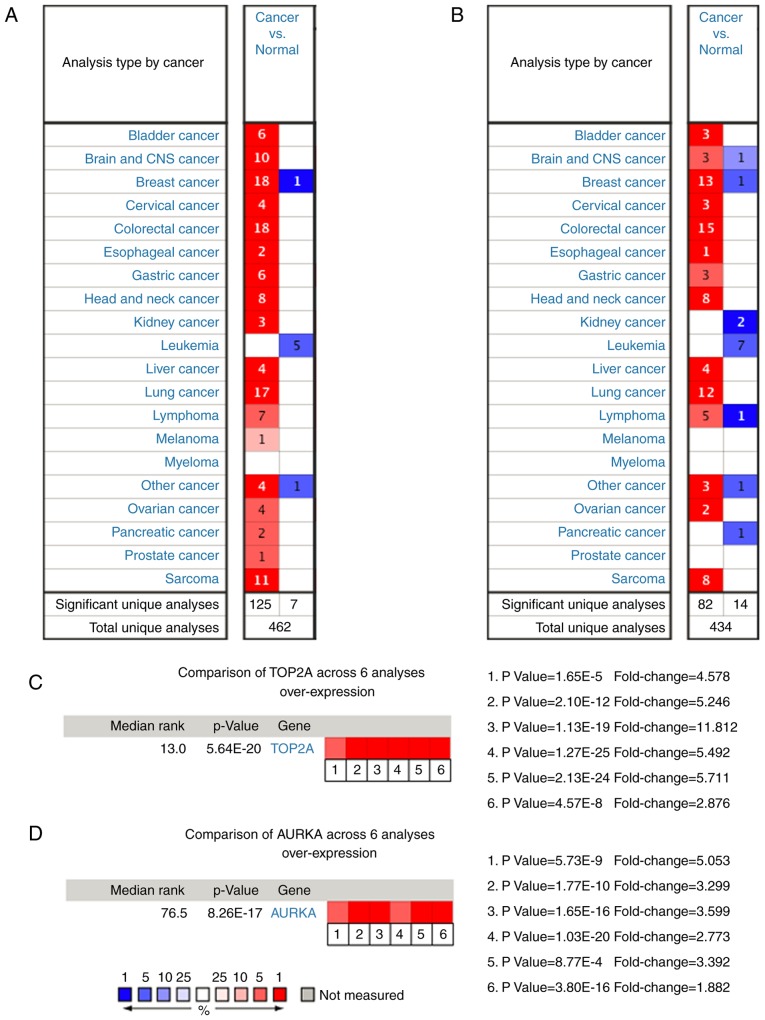
Figure 4.
Expression profiles of (A) TOP2A and (B) AURKA in 20 malignant tumor types are represented using the Oncomine database. The number represent the cases meeting the threshold for TOP2A and AURKA. Heat maps of (C) TOP2A and (D) AURKA gene expression in lung carcinoma samples vs. adjacent lung tissues in the Oncomine database. (C) 1. Lung carcinoma vs. normal lung, Beer et al (2002) (22). 2. Lung carcinoma vs. normal lung, Hou et al (2010) (23). 3. Lung carcinoma vs. normal lung, Landi et al (2008) (24). 4. Lung carcinoma vs. normal lung, Selamat et al (2012) (7). 5. Lung carcinoma vs. normal lung, Su et al (2007) (25). 6. Lung carcinoma vs. normal lung, Yamagata et al (2003) (26). (D) 1. Lung carcinoma vs. normal lung, Bhattacharjee et al (2001) (27). 2. Lung carcinoma vs. normal lung, Garber et al (2001) (28). 3. Lung carcinoma vs. normal lung, Hou et al (2010) (23). 4. Lung carcinoma vs. normal lung, Landi et al (2008) (24). 5. Lung carcinoma vs. normal lung, Selamat et al (2012) (7). 6. Lung carcinoma vs. normal lung, Su et al (2007) (25). The P-value for a gene is its P-value for the median-ranked analysis. The fold change represents the relative expression of the tumor tissue compared with the normal tissue. AURKA, aurora kinase A; TOP2A, DNA topoisomerase II α.