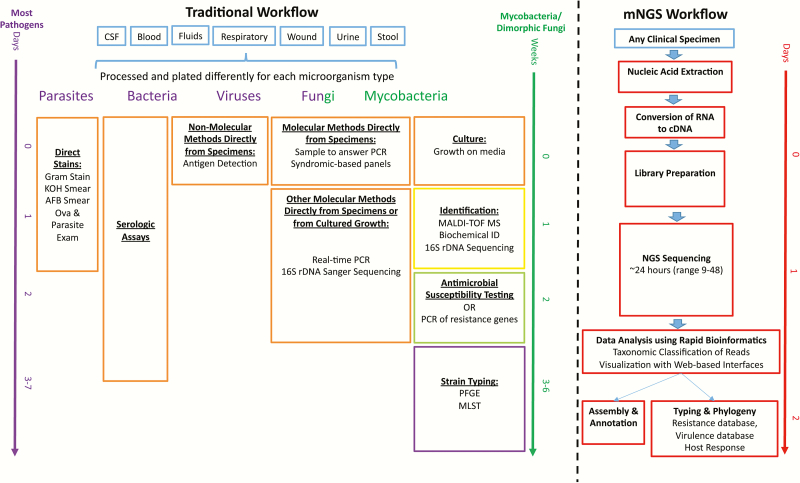
Figure 1.
Summary of the traditional timeline and workflow in diagnostic medical microbiology laboratories and the future state with the incorporation of metagenomic next-generation sequencing (mNGS) methodologies. Current organism detection techniques (orange), identification (yellow), antimicrobial susceptibility testing (green), and strain typing (purple) can take up to a week or longer from specimen collection (blue) to strain typing results. mNGS has the capability of greatly reducing turnaround times and providing all the data summarized in current methods in a single modality (red) and could potentially provide all of these within 24–48 hours of specimen receipt. To date, the available evidence is poor to use antibiotic resistance gene detection to predict phenotypic antimicrobial susceptibility testing profiles for clinical care [16]. Abbreviations: AFB, acid-fast bacilli; cDNA, complementary DNA; CSF, cerebrospinal fluid; KOH, potassium hydroxide; MALDI-TOF MS, matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry; MLST, multilocus sequence typing; mNGS, metagenomic next-generation sequencing; PCR, polymerase chain reaction; PFGE, pulsed-field gel electrophoresis; 16S rDNA, 16S ribosomal DNA.