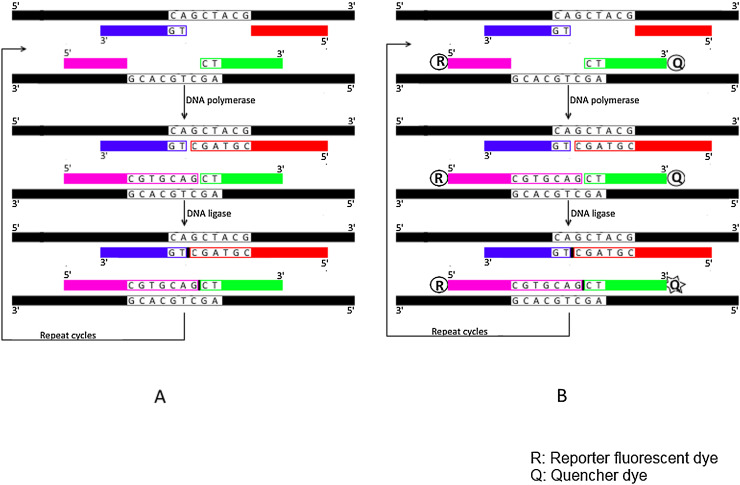
Fig. 2.
Schematic representation for Gap-LCR (GLCR) and Quantitative GLCR (QGLCR) techniques.
A) Gap-LCR (GLCR); Four oligonucleotide primers (each is colored/shaded differently) anneal perfectly with their complementary sequences of perfect match sample (colored in black) leaving a gap that is then filled by DNA polymerase and ultimately ligated (indicated by small black box) with DNA ligase enzyme to produce one ligated fragment for each template strand. Following repeated cycles, exponential ligation of products is achieved. B) Quantitative GLCR (QGLCR); Same as for GLCR except that the 5′ end of one primer is labeled with a reporter dye (R) while the 3′ end of the other primer that anneals adjacently on the same template strand is labeled with a quencher dye (Q). Following the two steps of polymerization and ligation, the ligated product (with small black box) brings reported dye (R) in close proximity with quencher dye (Q) resulting in fluorescence resonance energy transfer (FRET) from reporter dye into quencher dye upon excitation. Repeated cycles result in subsequent fluorescence of quencher dye.