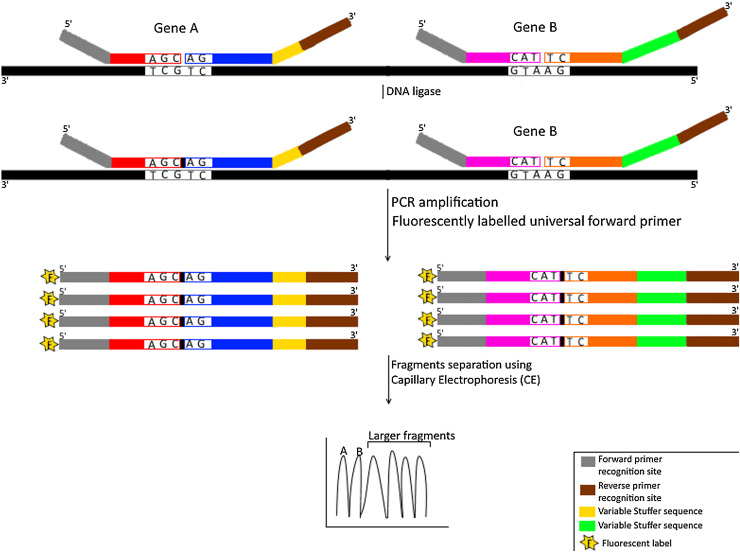
Fig. 4.
Schematic representation for Multiplex Ligation Dependent Probe Amplification (MLPA) technique.
The diagram shows simultaneous detection of 2 different genes (A & B) using four probes (two probes for each gene) that are colored/shaded differently. The first probe contains a recognition sequence for forward universal primer on its 5′ end and a specific sequence on its 3′ end that is complementary to template strand (colored in black). The second probe contains an allele specific sequence on its 5′ end to bind adjacently to the first probe on the same template strand and a reverse universal primer recognition sequence on its 3’end. A tailored stuffer sequence that is assigned for each target gene (for multiplexing purposes) is located between the reverse universal primer recognition sequence and the allele specific sequence of the second probe. DNA ligase seals the nick (indicated as small black box) between the two probes hybridizing to the same target gene. Fluorescently labeled universal forward primers are then used to generate 5′-fluorescently (indicated by letter F) labeled amplicons from ligated products. These products, with variable lengths attributed to the variable stuffer sequences used for each target gene, are separated using capillary electrophoresis.