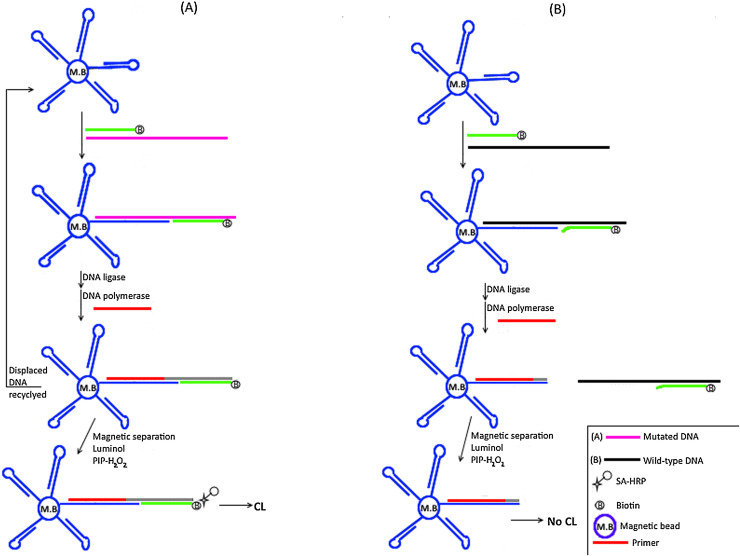
Fig. 9.
Schematic representation for Ligation-Mediated Strand Displacement Amplification Based Chemiluminescence Biosensor.
A) Mutated DNA; The hairpin probes that are immobilized on magnetic beads (indicated by letters M.B) hybridize with their complementary sequences on target DNA. A 5′ biotinylated (indicated by letter B) reporter probe anneals adjacently to the opened hairpin probe on the target DNA leaving a nick that is sealed with DNA ligase. Klenow fragment (exo−) polymerase and short primer are then used to synthesize a new strand that displaces target DNA. The latter initiates another cycle of hybridization with hairpin and biotinylated probes (indicated by letter B) for signal amplification. Following magnetic separation, horseradish peroxidase (HRP) conjugated streptavidin (indicated by star and small unfilled circle shapes) is added to bind biotinylated products for development of chemiluminescence signal through catalytic reaction of luminol- paraiodopenol-H2O2. B) Wild type DNA; Since there is no hybridization between target DNA and the hairpin probe, neither ligation with biotinylated reporter probe (indicated by letter B) nor displacement of the target DNA would occur and hence no development of chemiluminiscence signal.