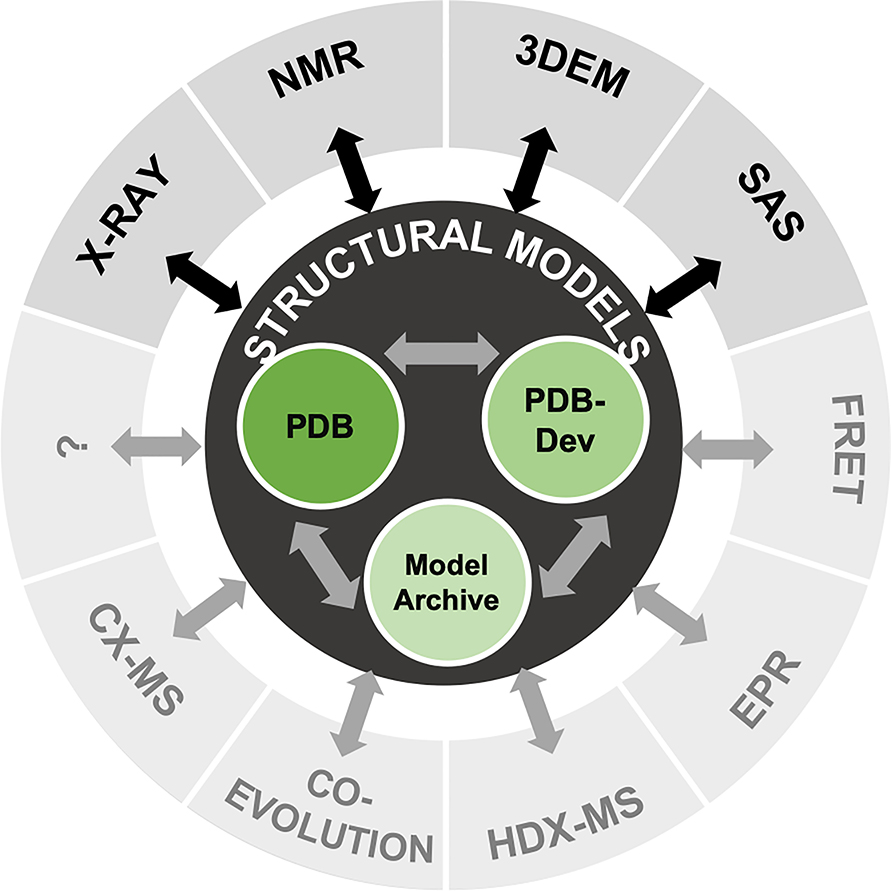
Figure 1. Illustration of federating structural models and experimental data.
At the center are the three structural biology model repositories: the PDB archive of experimentally determined structures of macromolecules (wwPDB consortium, 2019); the ModelArchive of in silico structural models (www.modelarchive.org); and the PDB-Dev prototype system for archiving integrative structures (Burley et al., 2017; Vallat et al., 2018). The outer circle indicates experimental data that contribute to integrative structural biology. Existing data exchange mechanisms for X-ray, NMR, 3DEM, and SAS data are represented by black arrows. Ongoing and future projects aim to develop methods for data exchange with archives for other types of experimental data as well as among the existing structural model repositories (gray arrows).