Fig. 2.
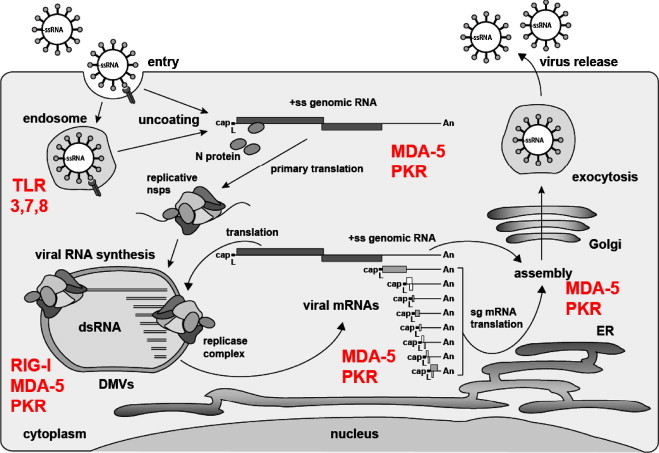
Coronavirus life cycle and RNA-specific pathogen recognition receptors. The coronavirus life cycle is illustrated together with PRRs with the potential to sense viral RNA. Coronaviruses enter their host cells either on the plasma membrane or via endosomes where they could be recognized by TLR 3, 7, or 8. Note that MHV is shown to be recognized by TLR7 in pDCs [92]. Upon uncoating the capped viral ssRNA is released into the host cell cytoplasm and could be sensed by MDA-5 or PKR due to secondary structures containing dsRNA domains. Viral RNA synthesis takes place in or at double membrane vesicles (DMVs) and involves the appearance of dsRNA [43], again potentially recognized by MDA-5 and PKR. The negative-sense RNAs arising as an intermediate of DMV-associated genome replication and transcription are possibly 5′-triphosphorylated and thus could be recognized by RIG-I. Finally, a nested set of viral mRNAs are released into the cytoplasm (putative sensors: MDA-5 or PKR) where they are translated. The full-length genomic RNA can also be translated and is eventually packaged into progeny virus particles which are released from the host cell via the exocytosis pathway.
