Fig. 1. The crystal structure of huIFN-α2 (YNS) triple mutant, designed to get a high affinity for IFNAR1 (23), shown in huIFN-α2-IFNAR ternary complex (A) and separately in 2 different projections (B, C).
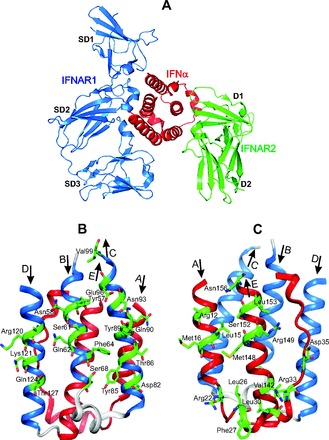
The colored blue B, C, and D α helices, which form the IFNAR1 binding site, and the side chains of residues, which contribute to the binding with IFNAR1 (23), are shown in front of B projection. The colored red A and E α helices and the AB loop, which form the IFNAR2 binding site (23), and the side chains of residues, which contribute to the binding with IFNAR2, are shown in front at C projection. The figure is reconstructed on the basis of the amino acid sequence, secondary structure, and coordinates of atoms from the Protein Data Bank under accession code 3SE3 (huIFN-α2-IFNAR ternary complex).
