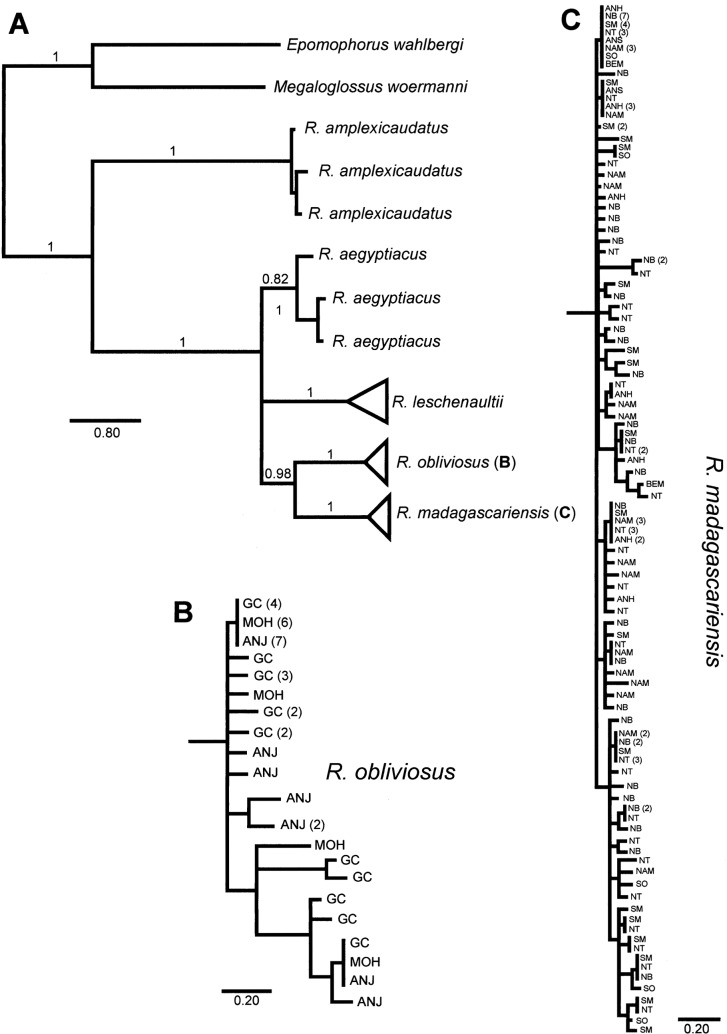
Fig. 2.
A) Bayesian consensus phylogram of Rousettus cytochrome b data. Branches of phylogram are labeled with posterior probability of bipartition, and scale bar for branches is provided below each tree. B) Detail of haplotype tree for samples of R. obliviosus. Tips of tree are labeled according to source islands: ANJ = Anjouan, MOH = Mohéli, and GC = Grande Comore. C) Detail of haplotype tree for samples of R. madagascariensis. Tips of tree are labeled according to source populations: ANH = Anjohibe, ANS = Anjanaharibe-Sud, NT = Northern Tip, NB = Nosy Be and Nosy Komba, SM = IIe Sainte Marie, BEM = Bemaraha/Ambohijanahary, NAM = Namoroka, and SO = Southern. Scale bar for branches is provided below each tree.