Figure 1. Identification of Targeted Dinucleotide Signatures Using DTECT.
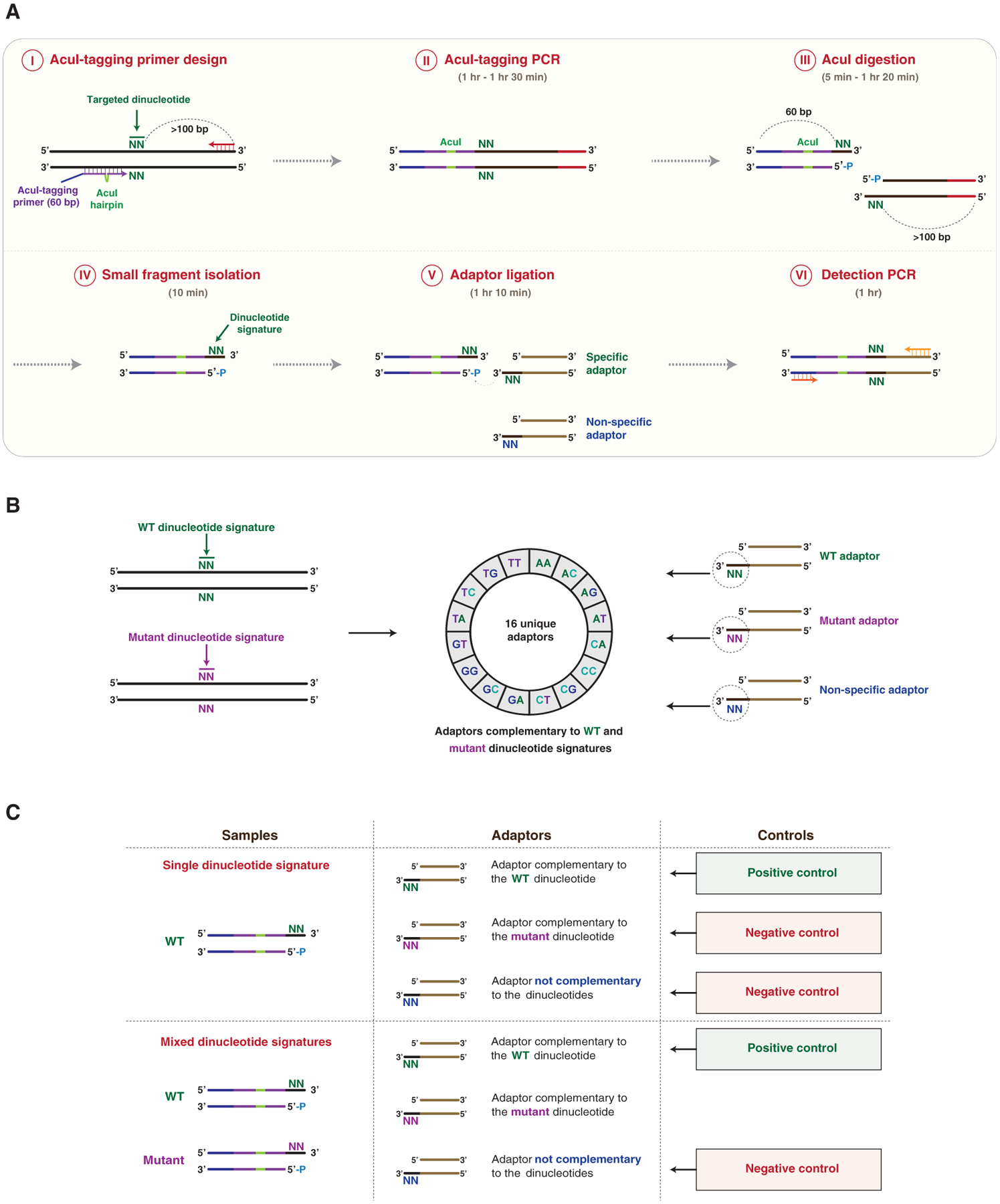
(A) Schematic representation of DTECT. The targeted genomic locus containing a hypothetical targeted dinucleotide (N = A, C, G, or T; green) is PCR amplified using a forward AcuI-tagging primer juxtaposed to the targeted dinucleotide and a locus-specific DNA primer (AcuI-tagging primer design and PCR, steps I and II). The AcuI-tagging primer (60 nt) consists of DNA sequences complementary to the genomic locus (purple) interrupted by a hairpin containing an AcuI recognition site (green), and a non-complementary DNA sequence (blue). The locus-specific reverse primer (red) is located >100 bp from the targeted dinucleotide. The obtained PCR product is subsequently cleaved by the AcuI restriction enzyme in a position adjacent to the targeted dinucleotide, resulting in the generation of two DNA fragments of 60 and >100 bp (AcuI digestion, step III). The 60 bp fragment containing the exposed signature of the targeted dinucleotide is then isolated using SPRI beads, with higher affinity toward >100 bp DNA products (small fragment isolation, step IV). The 60 bp fragment is then ligated to DNA adaptors containing 3′ overhangs of two bases complementary (specific) or not (non-specific) to the dinucleotide signature (adaptor ligation, step V). The ligated product is then subjected to PCR amplification for analytical or quantitative detection (detection PCR, step VI). The approximate time required for each step is indicated.
(B) Schematics of the DTECT adaptor library. Control (green) and mutant (purple) dinucleotide signatures (left panel) are detected using a library of 16 unique adaptors (middle panel). The library contains adaptors with dinucleotides complementary to the control (green) or mutant (purple) signature, as well as non-specific adaptors (blue) (right panel).
(C) Schematics of the positive and negative controls used in DTECT experiments to identify signatures of interest (e.g., mutant allele) in allele populations. In gDNA samples containing only the WT dinucleotide signature, the adaptor complementary to the WT dinucleotide signature (green) serves as a positive control, while the adaptor complementary to the mutant signature of interest (purple) and a non-specific adaptor (blue) are used as negative controls. In gDNA samples containing a mixture of the WT and the mutant dinucleotide signature, the adaptor complementary to the WT dinucleotide signature (green) is used as a positive control and a non-specific adaptor (blue) serves as a negative control. The adaptor complementary to the mutant dinucleotide signature (purple) is used to detect the presence of the variant of interest and quantify its frequency.
See also Figure S1.
