Figure 3. Detection and Quantification of Precision Genome Editing by CRISPR-Mediated HDR, Base Editing, and Prime Editing Using DTECT.
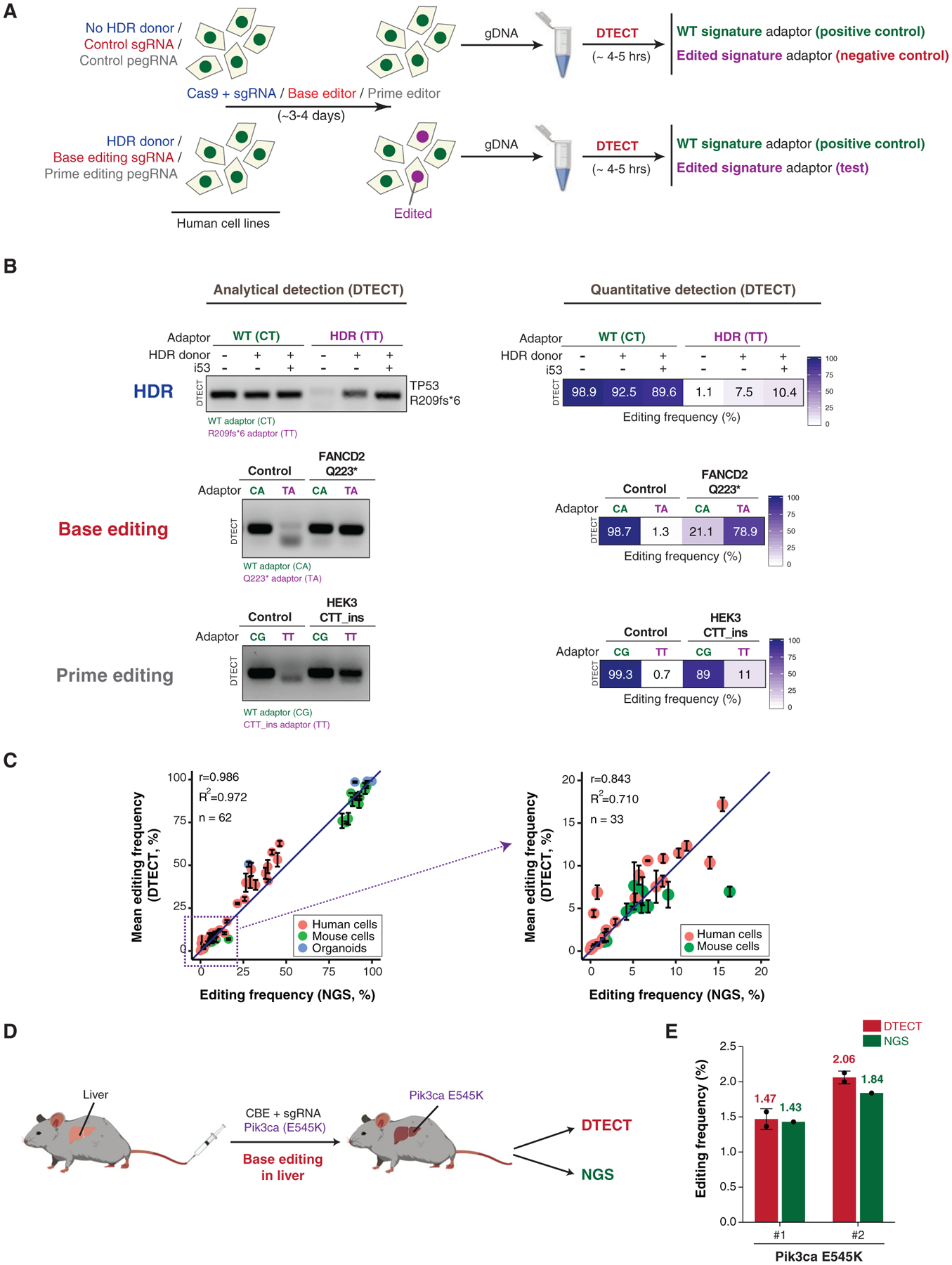
(A) Schematics of the protocol used to identify genomic changes introduced by CRISPR-dependent HDR, base editing, or prime editing. In HDR experiments (blue), HEK293T cells were transfected with Cas9 and an sgRNA targeting a gene of interest with or without donor DNA molecules. In base editing experiments (red), HEK293T cells were transfected with BE3 base editors with either control or base editing sgRNAs. Base editing experiments were also conducted in cells stably expressing FNLS-BE3. In prime editing experiments (gray), HEK293T cells were transfected with PE2 with or without pegRNA. gDNA was then extracted from cell populations and subjected to DTECT using adaptors specific for WT (green) or edited (purple) variants.
(B) Identification by DTECT of WT and HDR-edited (R209fs*6) TP53 alleles (top), WT and base-edited (Q223*) FANCD2 alleles (middle), and WT and prime-edited (CTT_ins) HEK3 alleles (bottom). Adaptors specific for the WT (CT, CA, and CG; green) or edited (TT and TA; purple) signatures were used in DTECT experiments. Captured samples were subjected to analytical PCR (left, 21 cycles) or qPCR (right). In the HDR experiment, cells were transfected with Cas9, sgRNA, and an ssODN specific for the TP53 locus with or without the HDR stimulatory factor i53. The ssODN was omitted in control reactions. In the base editing experiment, cells were transfected with BE3 and an sgRNA to induce Q223* in FANCD2. In prime editing experiments, cells were transfected with PE2 and pegRNA to introduce a CTT insertion in the HEK3 locus.
(C) Graphical representation of the correlation of DTECT- and NGS-based estimations of the frequency of genetic variants introduced by precision genome editing in human and mouse cells and mouse intestinal organoids (n = 62). Data points in the dashed box (frequency < 20%) of the left panel are enlarged in the right panel (n = 33). Error bars indicate the SEM of 2–5 independent replicates. The sources of the edited samples are indicated by distinct colors.
(D) Schematic representation of the experiments conducted to measure the efficiency of precision genome editing in vivo using DTECT. Editing of the mouse liver was performed by hydrodynamic injection of the cytidine base editor (CBE) FNLS-BE3 and an sgRNA to introduce the Pik3ca E545K variant. DTECT (red) and NGS (green) were used to determine the efficiency of editing in the mouse liver sample.
(E) Quantification by DTECT (red) and NGS (green) of the Pik3ca E545K variant introduced by CRISPR-mediated base editing in the mouse liver, as shown in (D). Error bars indicate the SD of 2 independent experiment. Dots represent individual data points.
See also Figures S4, S5, and S11.
