Fig 1.
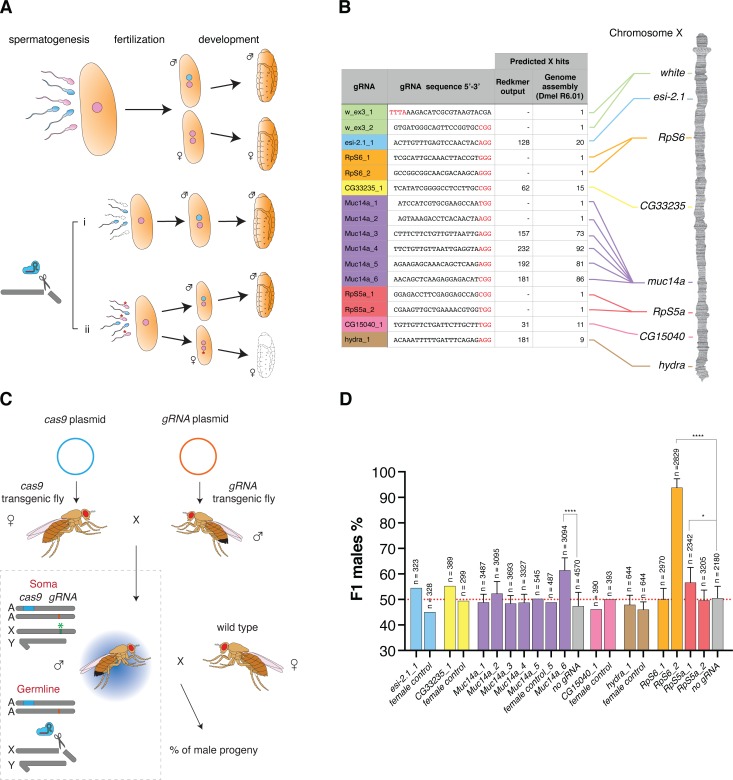
Development of sex ratio distorters in Drosophila (A) Model for pre-zygotic and post-zygotic effects on the reproductive sex-ratio. Compared to the unaltered process of development (top), an endonuclease targeting the X-chromosome during spermatogenesis (bottom) could either negatively affect the production or function of X-bearing sperm (i, prezygotic effect) or introduce genetic modifications that are detrimental to females inheriting such modified X-chromosomes during development (ii, postzygotic effect; e.g. the mutation of haploinsufficient genes). Pink: X-chromosome bearing gamete. Cyan: Y chromosome bearing gamete. The asterisk indicates gametes bearing modifications at the target site. (B) gRNA target sequences used in this study and their positions on the X-chromosome. The protospacer-adjacent motif (PAM) is indicated in red, and the X-chromosome hits predicted by Redkmer and the number of predicted perfect BLASTn hits to the X-chromosome in the Drosophila genome assembly (Dmel R6.01) are indicated. (C) Schematic of the experimental crosses. Females bearing the cas9 transgene were crossed to males carrying the gRNA transgene. Trans-heterozygote males were then crossed to wild-type females. Cas9 in concert with the gRNA cleaves the X-chromosome at the target site (green asterisk) in the germline and the effect is measured as the sex-ratio of the progeny. A: autosome; X: X-chromosome, Y; Y-chromosome. (D) Male sex-ratios in the offspring from crosses of βtub85D-cas9/gRNA with wild-type w females. Progeny of βtub85D-cas9/+ males crossed to wild type females (no gRNA) or from the reverse cross (βtub85D-cas9/gRNA females crossed to wild type males = female control) served as a control. Crosses were set as pools of males and females or as multiple male single crosses in which case error bars indicate the mean ± SD for a minimum of ten independent single crosses. For all crosses n indicates the total number of individuals (males + females) in the F1 progeny counted. P-values *p < 0.05, ****p < 0.0001. All crossing data can be found in S3 Table.