Fig 3.
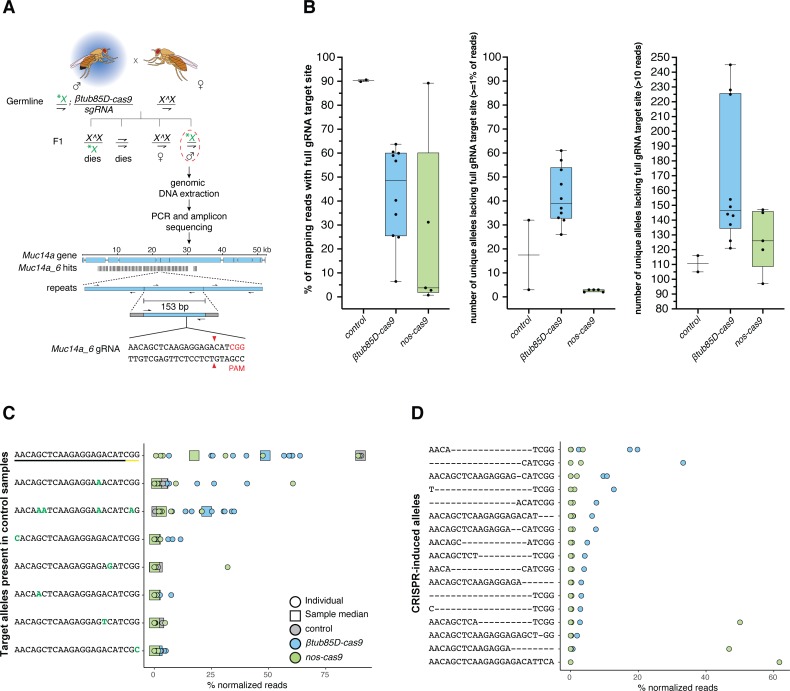
(A) Schematic of the genetic crosses to obtain shredded X-chromosomes from Muc14a_6/βtub85D-cas9 or Muc14a_6/nos-cas9 fathers for amplicon sequencing. Trans-heterozygous males for cas9 and the gRNA were crossed to females with attached-X-chromosomes (X^X). Offspring with supernumerary or lacking X-chromosomes are inviable leaving X^X/Y females and males carrying the X-shredded chromosome (patroclinous inheritance) which we selected and analysed. Genomic DNA from single males was extracted and a 153 bp DNA motif containing the Muc14a_6 gRNA target site was amplified with primers (thin arrows on repeats) containing Illumina Sequence adapters (grey boxes flanking the amplicon). As a comparison we used DNA from βtub85D-cas9/+ individuals lacking the gRNA. The shredded X-chromosome is indicated by a green asterisk. A dashed red circle indicates males selected for amplicon sequencing. Vertical black bars represent the number and location of the Muc14a_6 gRNA repeats within the Muc14a gene. (B) Analysis of allele variation at the Muc14a_6 target site by amplicon sequencing. Indicated is the percentage of all mapped reads that contain the complete, unaltered Muc14a_6 target site (left panel) in the control and experimental males. The middle and right panel show the number of reads harbouring all other unique alleles that represented at least 1% of all reads or were represented by 10 or more reads, respectively. (C) Analysis of unique alleles (including the wild type target site) at the Muc14a_6 target site that pre-existed in both control samples. Indicated are the relative frequencies of these alleles in each control and experimental male (circles) including the median frequency of each allele in all Muc14a_6/βtub85D-cas9 or Muc14a_6/nos-cas9 males (squares). (D) Analysis of de-novo alleles at the Muc14a_6 target site not present in control males and putative CRISPR-induced alleles. In C and D we considered only alleles that represented ≥1% of normalized reads in at least one male sample.