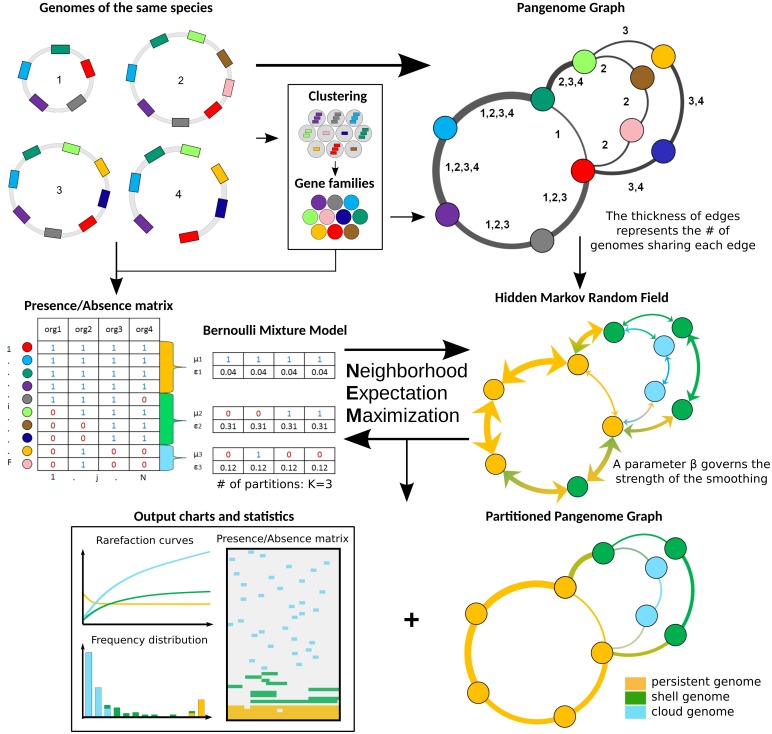
Fig 1. Flowchart of PPanGGOLiN on a toy example of 4 genomes.
The method requires annotated genomes of the same species with their genes clustered into homologous gene families. Annotations and gene families can be predicted by PPanGGOLiN or directly provided by the user. Based on these inputs, a pangenome graph is built by merging homologous genes and their genomic links. Nodes represent gene families and edges represent genomic neighborhood. The edges are labeled by identifiers of genomes sharing the same gene neighborhood. In parallel, gene families are encoded as a presence/absence matrix that indicates for each family whether or not it is present in the genomes. The pangenome is then divided into K partitions (K = 3 in this example) by estimating the best partitioning parameters through an Expectation-Maximization algorithm. The method involves the maximization of the likelihood of a multivariate Bernoulli Mixture Model taking into account the constraint of a Markov Random Field (MRF). The MRF network is given by the pangenome graph and it favors two neighbors to be more likely classified in the same partition. At the end of this iterative process, PPanGGOLiN returns a partitioned pangenome graph where persistent, shell and cloud partitions are overlaid on the neighborhood graph. In addition, many tables, charts and statistics are provided by the software. The number of partitions (K) can either be provided by the user or determined by the algorithm.