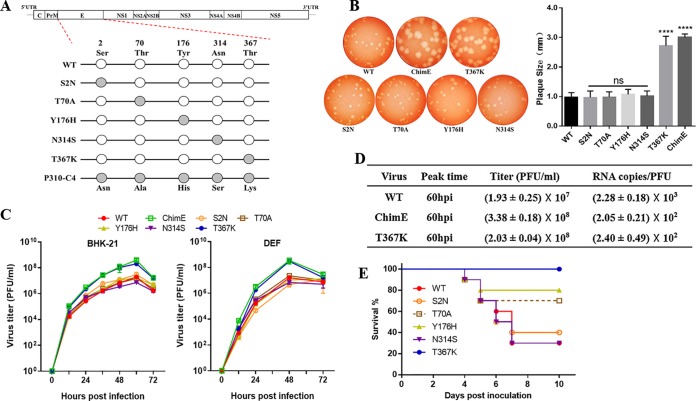
FIG 2.
Characteristics of viruses harboring E protein site-directed mutations in vitro and in vivo. (A) Schematics of the construction of five individual E protein site-directed mutant viruses based on the infectious WT clone. Site-directed mutations at E2, E70, E176, E314, and E367 were generated and constructed by fusion PCR. (B) Plaque phenotypes of the WT, ChimE, and E protein site-directed mutant viruses. The virus-infected BHK-21 monolayers were incubated under an overlay medium containing 1% agarose and then stained with neutral red at 3 dpi (left). Twenty plaques for each virus were chosen at random to calculate plaque sizes (right). The data are presented as means and standard deviation (SD) and were tested for statistical significance using the Student’s t test. ns, no significant difference; ****, P < 0.0001. (C) Virus growth kinetics. BHK-21 and DEF cells were infected with WT, ChimE, and E protein site-directed mutant viruses at an MOI of 0.01. At each indicated time point, the extracellular infectious viruses were quantified using a plaque assay. The data represent the means and SD of three independent cell cultures. The result is shown as a representative of three independent experiments. (D) Virus titer and RNA copy number/PFU ratios. From 12 to 96 hpi, supernatants were collected at an interval of 12 h. Infectious virions were determined using a plaque assay, and the copies of viral RNA at the peak titer time were quantified by qRT-PCR. (E) Survival curve for the groups of 4-day-old ducklings (n = 10 ducks per group) s.c. infected with the indicated viruses (1 × 105 PFU/bird).