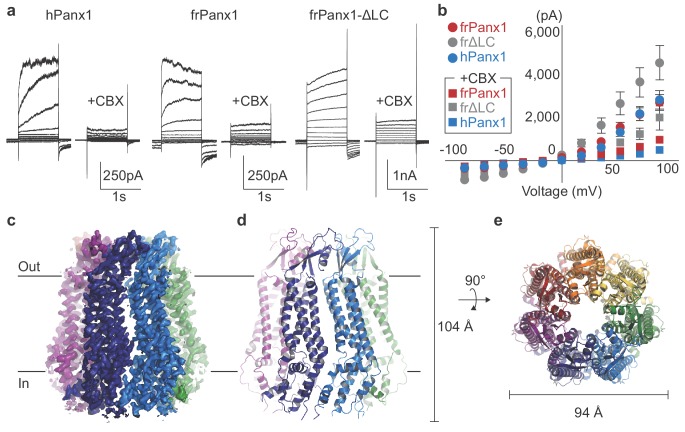
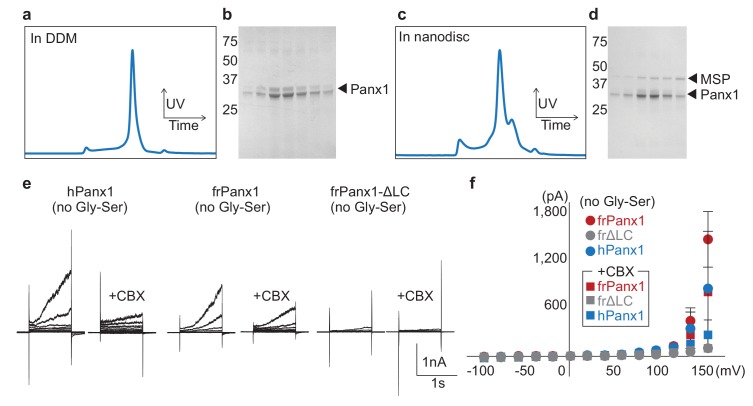
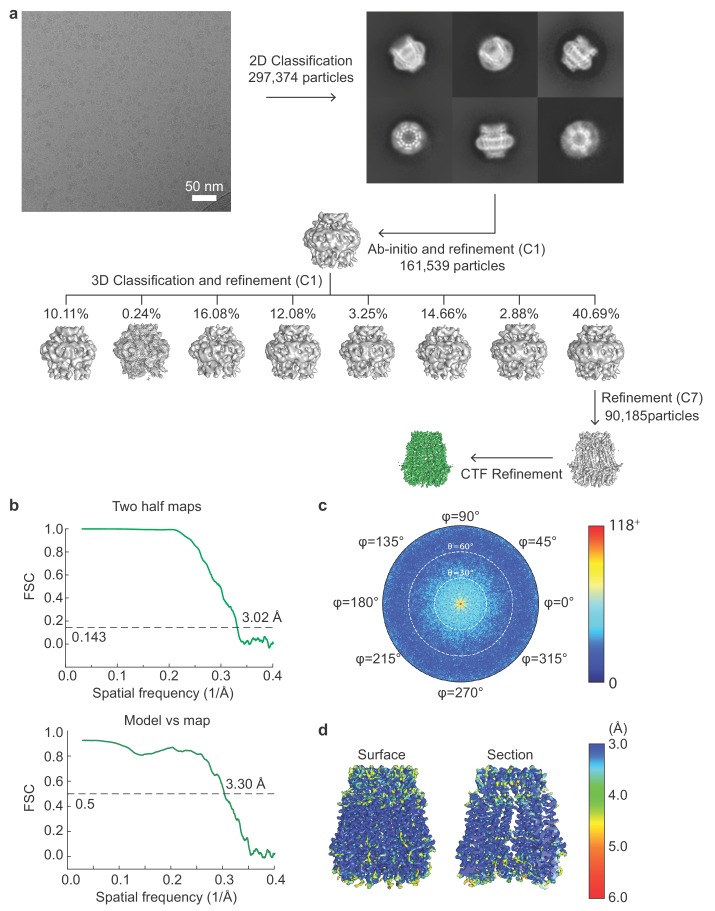
Figure 1. frPanx1 forms a heptameric ion channel.
(a) Whole-cell patch clamp recordings from HEK 293 cells expressing hPanx1, frPanx1, and frPanx1-ΔLC. Cells were clamped at −60 mV and stepped from −100 mV to +100 mV for 1 s in 20 mV increments. To facilitate electrophysiological studies, we inserted a Gly-Ser motif immediately after the start Met to enhance Panx1 channel opening as we previously described (Michalski et al., 2018). CBX (100 μM) was applied through a rapid solution exchanger. (b) Current-voltage plot of the same channels shown in (a). Recordings performed in normal external buffer are shown as circles, and those performed during CBX (100 μM) application are shown as squares. Each point represents the mean of at least three different recordings, and error bars represent the SEM. (c) EM map of frPanx1-ΔLC shown from within the plane of the membrane. Each protomer is colored differently, with the extracellular side designated as ‘out’ and the intracellular side as ‘in.’ (d) Overall structure of frPanx1-ΔLC viewed from within the lipid bilayer. (e) Structure of frPanx1 viewed from the extracellular face.