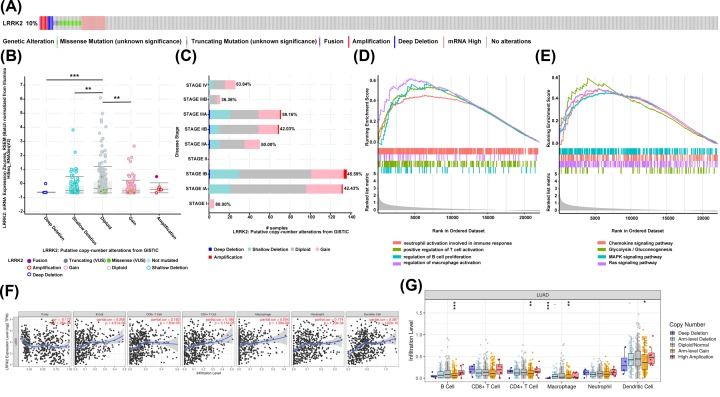
Figure 8. Further exploration of LRRK2.
(A) OncoPrint of LRRK2 alterations in LUAD cohort. The different types of genetic alterations are highlighted in different colors. (B) LRRK2 expression in different LRRK2 CNV groups. **, P<0.01, ***, P<0.001 (C) Distribution of LRRK2 CNV frequency in different stage subgroups. The percentage number on the right of the bar indicates the ratio of patients with LRRK2 deep deletion, shallow deletion, and gain in all this subgroup patients. (D and E) Significantly enriched GO terms and KEGG pathways of LRRK2 by GSEA (P-value < 0.05). (F) LRRK2 expression is significantly related to tumor purity and has significant positive correlations with infiltrating levels of B cell, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in LUAD. (G) LRRK2 CNV affects the infiltrating levels of B cells, CD4+ T CELL, macrophages, and dendritic cells in LUAD. *, P<0.05, **, P<0.01, ***, P<0.001.