Figure 2.
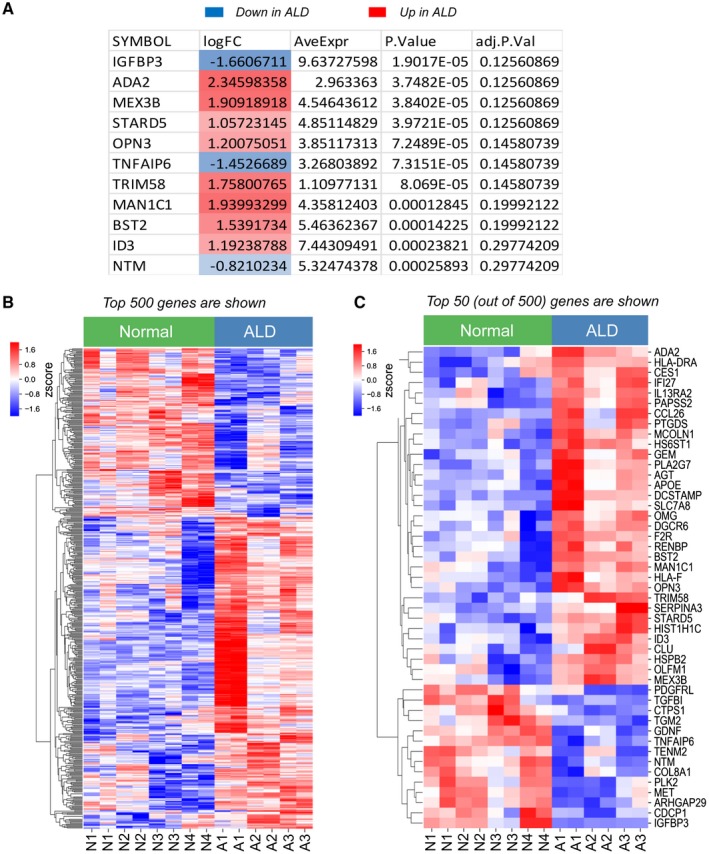
Eleven most dysregulated genes that best discriminate ALD hHSCs from normal hHSCs. (A) Eleven genes that are significantly differentially expressed (red, up‐regulated; blue, down‐regulated) between ALD and normal hHSCs. (B,C) Heatmap showing the relative gene expression between the top 500 (B) or top 50 (C) most differentially expressed genes in normal and ALD hHSCs. The genes were grouped by similarity to each other using hierarchical clustering. The Benjamini‐Hochberg method for multiple test corrections was applied to differentiate the expression results. Eleven genes were changed significantly with the Benjamini‐Hochberg method (adjusted P < 0.3). Abbreviations: adj.P.Val, multiple‐test‐corrected P value using the method of Benjamini‐Hochberg; AveExpr, average expression across all samples; logFC, log fold change in expression between ALD and normal hHSCs; P.Value, P value for the comparison. Abbreviations: ADA2, adenosine deaminase 2; AGT, angiotensinogen; APOE, apolipoprotein E; ARHGAP29, rho GTPase activating protein 29; BST2, bone marrow stromal cell antigen 2; CCL26, C‐C motif chemokine ligand 26; CDCP1, CUB domain containing protein 1; CES1, carboxylesterase 1; CLU, clusterin; COL8A1, collagen type VIII alpha 1 chain; CTPS1, CTP synthase 1; DCSTAMP, dendrocyte expressed seven transmembrane protein; DGCR6, DiGeorge syndrome critical region gene 6; F2R, coagulation factor II thrombin receptor; GDNF, glial cell derived neurotrophic factor; GEM, GTP binding protein overexpressed in skeletal muscle; HIST1H1C, H1.2 linker histone, cluster member; HLA‐DRA, major histocompatibility complex, class II, DR alpha; HLA‐F, major histocompatibility complex, class I, F; HS6ST1, heparan sulfate 6‐O‐sulfotransferase 1; HSPB2, heat shock protein family B (small) member 2; ID3, inhibitor of DNA binding 3; IFI27, interferon alpha inducible protein 27; IGFBP3, insulin like growth factor binding protein 3; IL13RA2, interleukin 13 receptor subunit alpha 2; MAN1C1, mannosidase alpha class 1C member 1; MCOLN1, mucolipin 1; MET, MET proto‐oncogene, receptor tyrosine kinase; MEX3B, mex‐3 RNA binding family member B; NTM, neurotrimin; OLFM1, olfactomedin 1; OMG, oligodendrocyte myelin glycoprotein; OPN3, opsin 3; PAPSS2, 3,‐phosphoadenosine 5,‐phosphosulfate synthase 2; PDGFRL, platelet derived growth factor receptor like; PLA2G7, phospholipase A2 group VII; PLK2, polo like kinase 2; PTGDS, prostaglandin D2 synthase; RENBP, renin binding protein; SERPINA3, serpin family a member 3; SLC7A8, solute carrier family 7 member 8; STARD5, StAR related lipid transfer domain containing 5; TENM2, teneurin transmembrane protein 2; TGFBI, transforming growth factor beta induced; TGM2, transglutaminase 2; TNFAIP6, tumor necrosis factor‐alpha‐induced protein; TRIM58, Tripartite motif containing 58.
