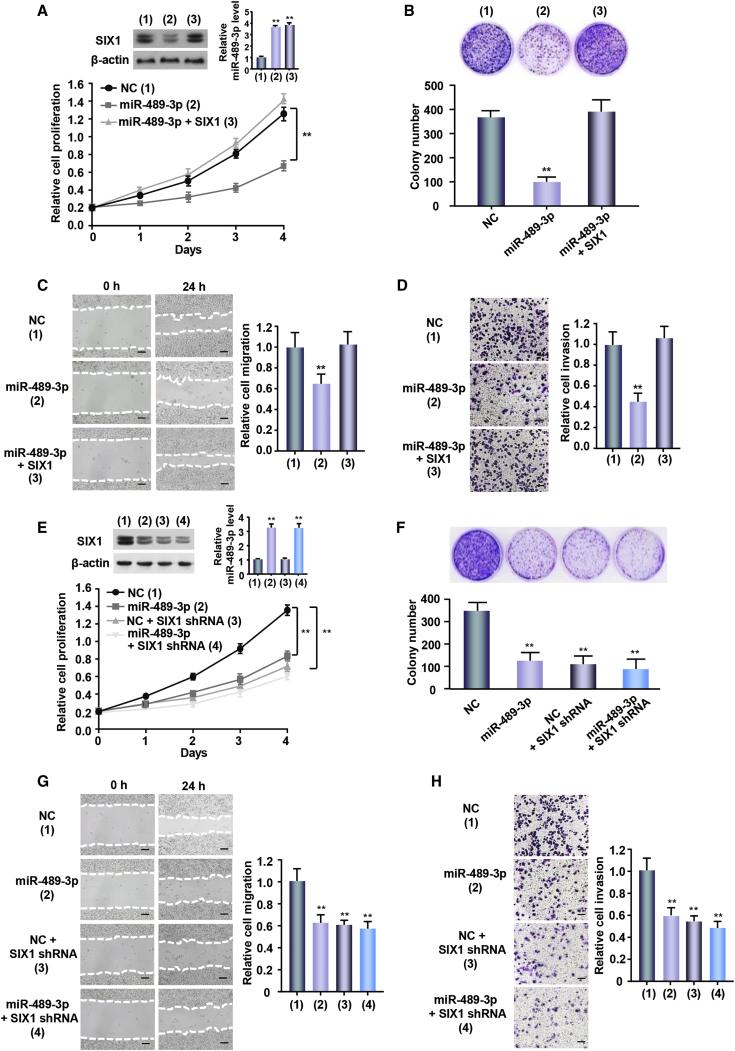
Figure 2.
miR-489-3p/SIX1 Axis Regulates Proliferation, Migration, and Invasion in Melanoma Cells
(A) A CCK-8 assay was performed to detect relative proliferation compared to day 0 of the indicated groups of cells. The immunoblot shows SIX1 expression. Bar graphs indicate miR-489-3p expression performed by qRT-PCR. (B) A colony formation assay was performed in A375 cells transfected as in (A). Representative graphs show colonies in plates. Bar graphs show colony number. (C and D) Wound healing and Transwell invasion assays were performed in A375 cells transfected as in (A). Bar graphs show relative cell migration and invasion. (E-F) Lentivirus-mediated SIX1 knockdown (SIX1 shRNA) or control A375 cells were transfected with NC or miR-489-3p mimics. CCK-8 assay (E) and colony formation (F) were performed and analyzed. The immunoblot shows SIX1 expression. Bar graphs indicate miR-489-3p expression performed by qRT-PCR. (G-H) Lentivirus-mediated SIX1 knockdown (SIX1 shRNA) or control A375 cells were transfected with NC or miR-489-3p mimics. Wound healing (G) and Transwell invasion assays (H) were performed. Bar graphs show relative cell migration and invasion. Scale bars, 100 μm. (*p < 0.05, **p < 0.01 versus corresponding control). Each experiment was repeated at least twice and one representative result is given. The data shown are the average values with error bars representing the SE (A–H).