Figure 3.
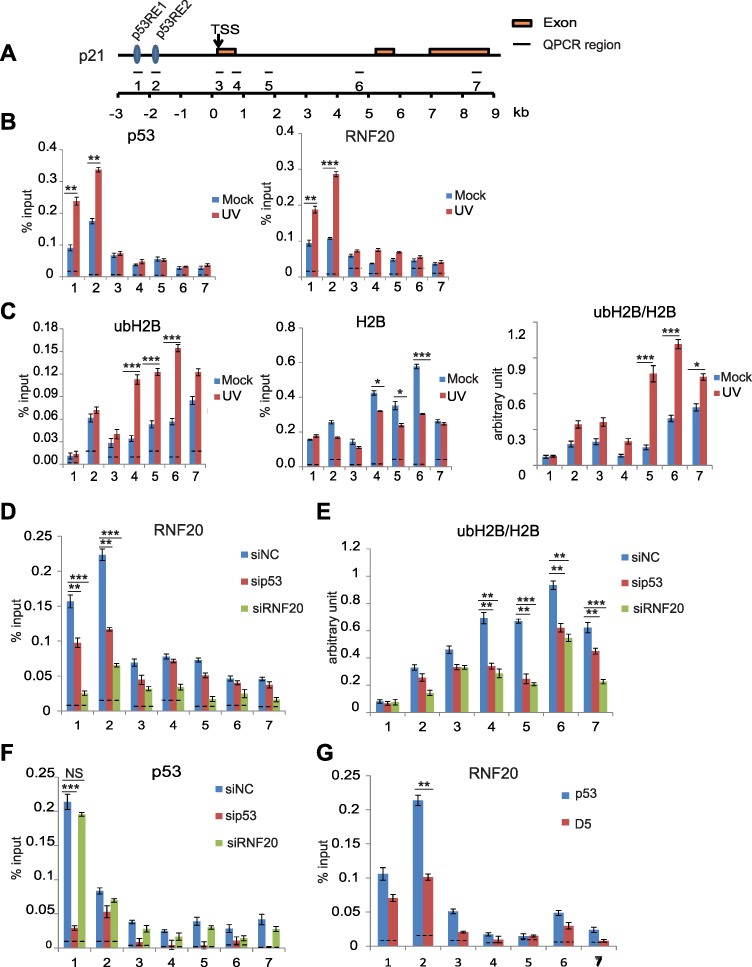
p53 mediates the recruitment of the RNF20/40 complex to the promoter region of p21. (A) Schematic representation of p21 genomic locus. p53 RE1, p53 response element 1; p53 RE2, p53 response element 2; TSS, transcription start site. (B) DNA damage induces p53 and RNF20 to the p21 gene promoter region. HCT116 cells were treated with or without UV-C (20 J/m2). After 8 h recovery, ChIP analyses on the p21 locus were performed using anti-p53 or anti-RNF20 antibodies. An irrelevant IgG was used for a control shown as the dotted lines. Quantitative PCR amplification regions are indicated in the schematic diagram. The sequence of each primer is shown in Supplementary Table S3. (C) ubH2B was enriched at the p21 gene body region in response to DNA damage. ChIP analyses were performed with anti-ubH2B and anti-H2B antibodies. The relative enrichment of ubH2B was examined by the relative ratio of ubH2B vs. H2B. (D) p53 mediates the recruitment of RNF20 at the promoter region of p21. HCT116 cells were transfected with control siRNA (siNC), p53 siRNA, and RNF20 siRNA, respectively, following UV-C (20 J/m2). ChIP analyses on the p21 locus were performed. The recruitment of RNF20 was examined by anti-RNF20 antibodies in ChIP assays. (E) Both p53 and RNF20 regulate ubH2B at the gene body region of p21. Knockdown of p53 or RNF20 was achieved by siRNA treatment. The relative ratio of ubH2B vs. H2B was examined by ChIP assays with anti-ubH2B and anti-H2B antibodies. (F) RNF20 is not required for the recruitment of p53. Following the siRNA treatment to knockdown p53 or RNF20, the enrichment of p53 was examined by ChIP assay with anti-p53 antibodies. (G) The CTD of p53 is required for the recruitment of RNF20. Full-length p53 and the D5 mutant were expressed in HCT116 p53−/− cells. ChIP analyses on RNF20 were performed. Data are represented as mean ± standard deviation (SD) as indicated from three independent experiments. Significance of differences was evaluated by Student’s t-test. NS, nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001.
