Figure 3.
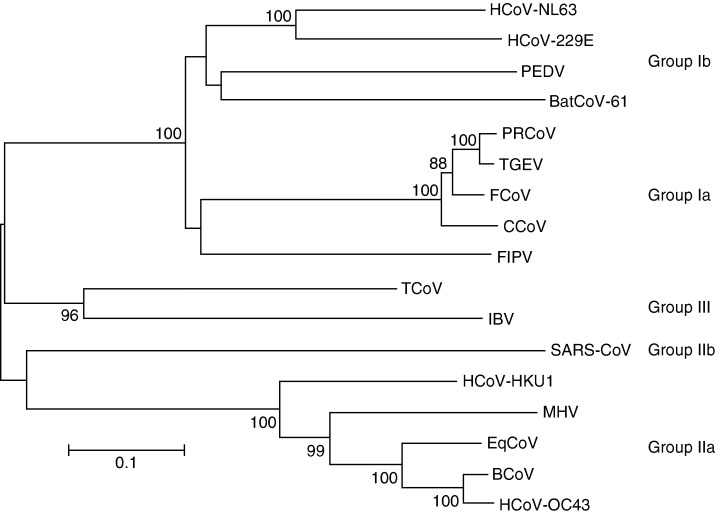
Phylogenetic analysis of coronaviruses. Analysis was carried out with the nucleotide sequences of a part of the spike gene (HCoV‐NL63 position 22854–24319) using the neighbour‐joining method of the mega program. Bootstrap resampling (1000 replicates) was employed to place approximate confidence limits on individual branches; bootstrap values above 80 are shown. The GenBank accession number of the sequences used in this phylogenetic analysis are HCoV‐229E: n; CCoV (canine coronavirus) n; FIPV (feline infectious peritonitis virus) n; PEDV (porcine epidemic diarrhea virus) n; TGEV (transmissible gastroenteritis virus): n; PRCoV (porcine respiratory coronavirus) n; MHV (mouse hepatitis virus) n; BCoV (bovine coronavirus) n; HCoV‐OC43: n; SARS‐CoV: n; IBV (avian infectious bronchitis virus) n; TCoV (turkey coronavirus) n; BatCoV strain61: n and HCoV‐HKU1: n.
