Figure EV1. The Sen1 NIM is conserved in close yeast species.
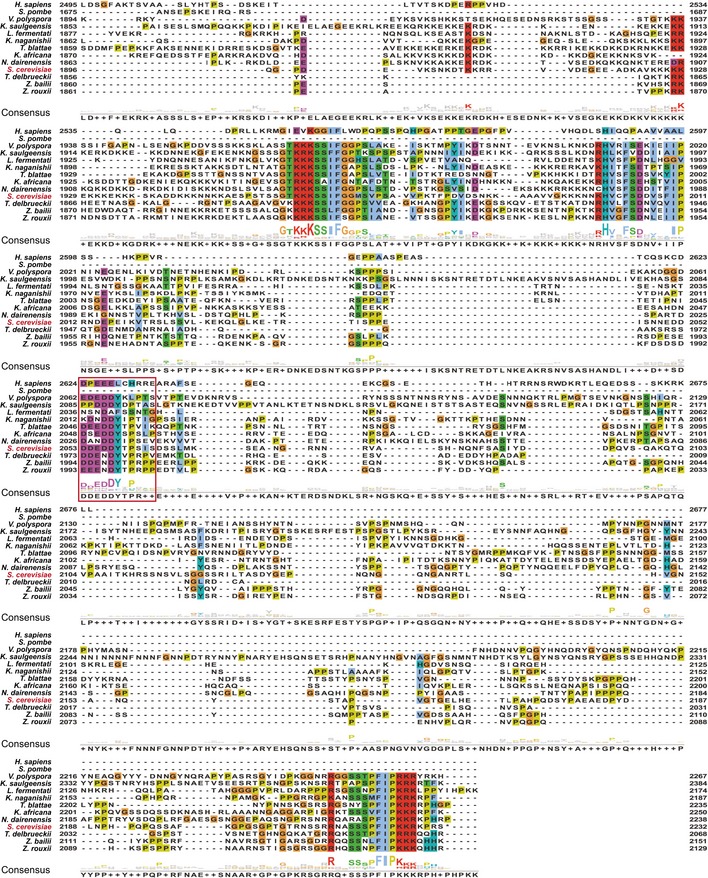
Saccharomyces cerevisiae Sen1 protein sequence was submitted to blastp excluding the genus Saccharomyces from the search. The ten most conserved protein sequences together with Sen1 orthologues from Homo sapiens (SETX) and Schizosaccharomyces pombe were aligned to S. cerevisiae Sen1 using clustal omega. Visualization of the alignment and calculation of the consensus sequences ware performed with Jalview (Waterhouse et al, 2009). Amino acids are coloured according to clustal colour scheme. The protein identifiers for the proteins used in the alignment are the following: Q7Z333 (H. sapiens), Q92355 (S. pombe), XP_001644478.1 (Vanderwaltozyma polyspora), SMN21961.1 (Kazachstania saulgeensis), SCV99407.1 (Lachancea fermentati), CCK69072.1 (Kazachstania naganishii), XP_004181671.1 (Tetrapisispora blattae), XP_003955130.1 (Kazachstania Africana), XP_003672383.1 (Naumovozyma dairenensis), Q00416 (S. cerevisiae), XP_003680903.1 (Torulaspora delbrueckii), CDF91445.1 (Zygosaccharomyces bailii) and GAV52597.1 (Zygosaccharomyces rouxii). The NIM is indicated by a red box.
