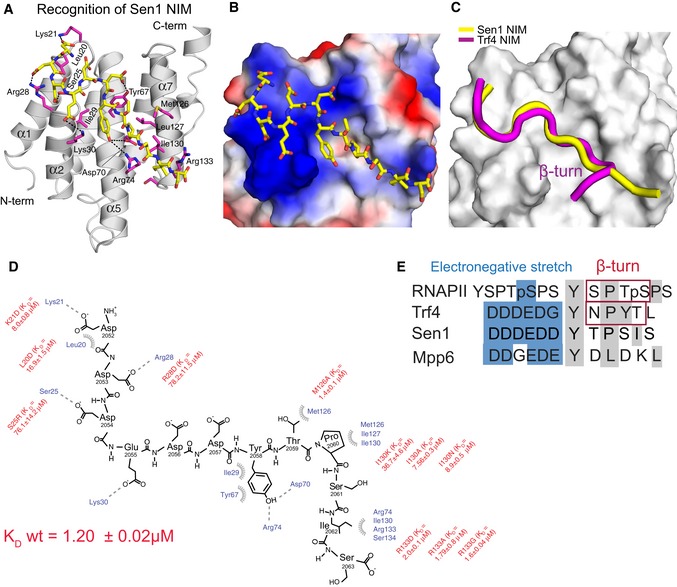
Figure 2. Recognition of Sen1 NIM by Nrd1 CID .
-
ANMR structure of Nrd1 CID bound to Sen1 NIM. The NIM peptide is represented in yellow sticks (only non‐hydrogen atoms are shown), and Nrd1 CID is shown as a grey ribbon model. Nrd1 CID residues that form hydrophobic contacts and putative hydrogen bonds to Sen1 NIM peptide are shown in magenta sticks.
-
BElectrostatic surface representation of Nrd1 CID (electropositive in blue; electronegative in red; neutral in white) with the Sen1 NIM peptide (represented in yellow sticks; only non‐hydrogen atoms are shown). The upstream electronegative stretch of Sen1 NIM interacts with an electropositive pocket of Nrd1 CID, while the C‐terminal part of the peptide adopts an extended conformation that docks in a hydrophobic pocket of Nrd1 CID.
-
CSuperposition of Nrd1 CID–Sen1 NIM (yellow) and Nrd1 CID–Trf4 NIM (magenta) complexes, displaying only peptide ribbons on the surface of Nrd1 CID. The comparison highlights the extended and β‐turn conformations of Sen1 NIM and Trf4 NIM, respectively.
-
DScheme showing contacts (putative H‐bonds and hydrophobic contacts) and energetics between the Sen1 NIM peptide and Nrd1 CID. Equilibrium binding experiments with the protein mutants were monitored by FA. L20D mutant disrupts the hydrophobic contact with F17 and impairs the overall geometry of the α1‐α2 loop that contributes to the interaction with the upstream electronegative stretch of NIM. K D (wild‐type Nrd1 CID‐Sen1 NIM) equals 1.20 ± 0.02 μM.
-
EAlignment of RNAPII CTD and CTD mimics (Trf4 NIM, Sen1 NIM and Mpp6 CTD mimic). Blue and grey boxes highlight the upstream electronegative stretches and hydrophobic regions, respectively. Previous structural works reported that S/NPXX motifs form the β‐turn conformation (Kubicek et al, 2012; Tudek et al, 2014).
