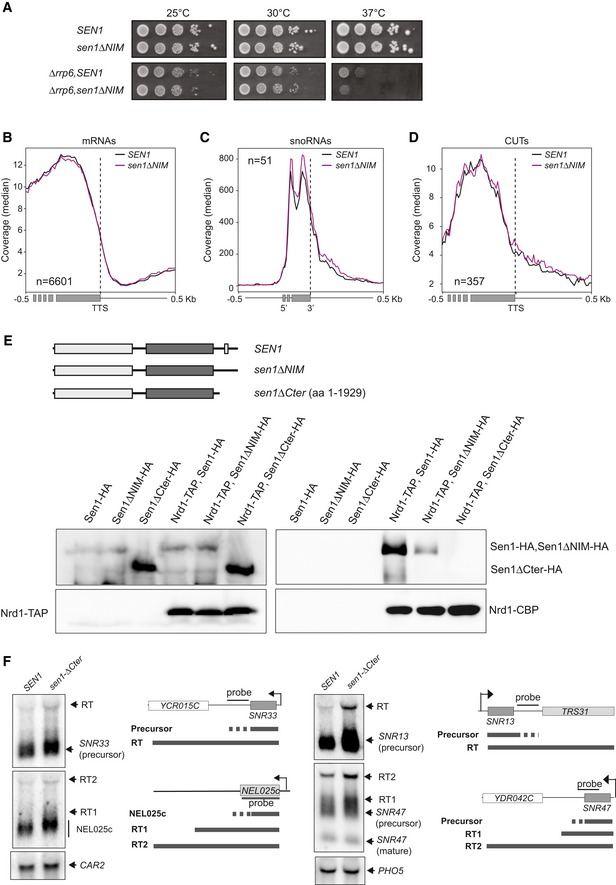
Figure EV3. The interaction of Sen1 with Nrd1 and Nab3 is not essential for non‐coding transcription termination (related to Fig 3).
-
AGrowth tests of the sen1∆NIM mutant in either a wt or a ∆rrp6 background.
-
B–DMetagene analyses of RNAseq experiments performed in a ∆rrp6 background in the presence of the wt or the ∆NIM version of Sen1. The profile corresponds to the median coverage (reads per 107 reads mapping at each genomic position) from 0.5 kb upstream to 0.5 kb downstream of the annotated transcription termination site (TTS) of protein‐coding genes (B) and CUTs (D) or the 3′ end of the mature snoRNAs (C). Experiments were performed in biological duplicates.
-
EDeletion of Sen1 C‐terminal domain completely abolishes the interaction of Sen1 with Nrd1. Top: scheme of proteins analysed in these experiments. Bottom: CoIP assays using Nrd1‐TAP as the bait. Representative gel of one out of two independent experiments. Antibodies used for protein detection are detailed in Appendix Table S3.
-
FDeletion of Sen1 Cter provokes minor transcription termination defects at typical NNS‐dependent non‐coding genes. Northern blot assays performed in a ∆rrp6 background. Results correspond to one out of two independent biological replicates. The CAR2 and PHO5 RNAs are detected as a loading controls. Probes used for RNA detection are described in Appendix Table S6.
