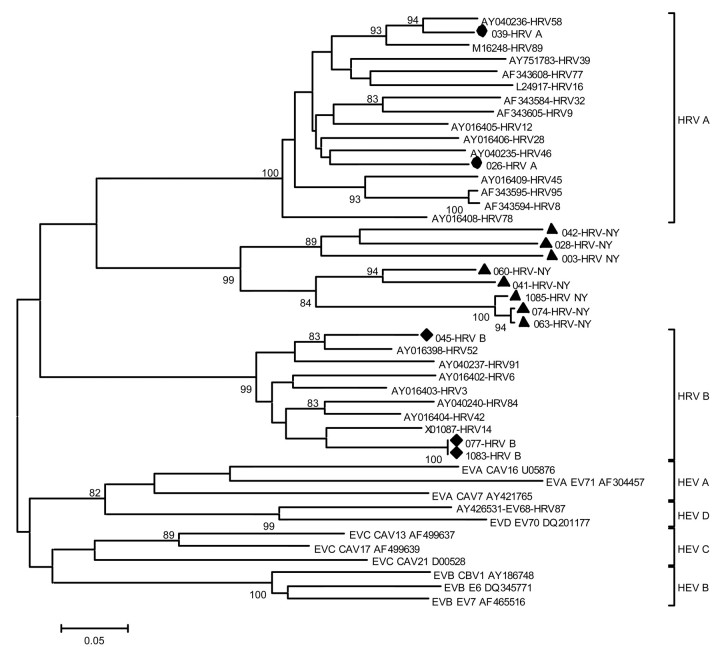
Figure 1.
Dendrogram of isolates of rhinovirus from New York State (NYS) and of selected reference isolates of enterovirus and rhinovirus. The nucleotide sequence of VP4 was used to reconstruct a phylogenetic tree by use of the neighbor-joining method applying a Kimura 2-parameter model. Isolates from NYS that are related to human rhinovirus (HRV) group A (DQ875920 and DQ875923 [•]; DQ875923 and DQ875920) and HRV group B (DQ875922, DQ875927, and DQ875930 [♦]) are shown, as are isolates (DQ875921, DQ875924, DQ875925, DQ875926, DQ875928, DQ875929, DQ875931, and DQ875932 [▴]) that cluster as a distinct genetic clade, HRV-NY. The scale bar indicates nucleotide substitutions per site, and bootstrap values (percentage of 1000 pseudoreplicates) are shown at relevant branches