Fig. 1.
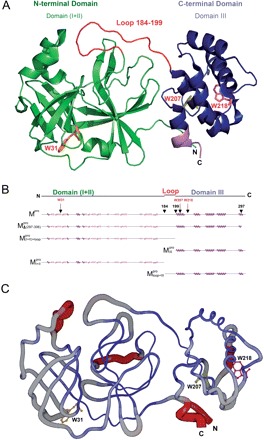
Structural features of the SARS-CoV Mpro. (A) The three structural domains of SARS-CoV Mpro (pdb code: 1uk3) are shown in ribbon representation. The N-terminal domain (residues 1–183) is shown in green, C-terminal domain (residues 200–306) in blue, the loop (residues 184–199) connecting the two domains in red, the deletion region (residues 297–306) of 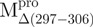 in violet. The tryptophanyl residues Trp31, Trp207 and Trp218 are shown in orange, yellow and pink, respectively, as bond model. Generate with Pymol. (B) Secondary structure features of the enzyme and the truncated regions of the various truncation mutants. (C) The structural flexibility of the enzyme is shown as a variable worm of the polypeptide chain based on the B factor of the pdb file (pdb code: 1uk3). The rigid structure is shown as a thinner worm in blue. The more flexible peripheral regions are shown in red. Only one of the subunits is shown. This figure was generated with SPOCK (http://quorum.tamu.edu).
in violet. The tryptophanyl residues Trp31, Trp207 and Trp218 are shown in orange, yellow and pink, respectively, as bond model. Generate with Pymol. (B) Secondary structure features of the enzyme and the truncated regions of the various truncation mutants. (C) The structural flexibility of the enzyme is shown as a variable worm of the polypeptide chain based on the B factor of the pdb file (pdb code: 1uk3). The rigid structure is shown as a thinner worm in blue. The more flexible peripheral regions are shown in red. Only one of the subunits is shown. This figure was generated with SPOCK (http://quorum.tamu.edu).
