Figure 4.
Two M48USP Histidines Can Participate in the Catalytic Triad
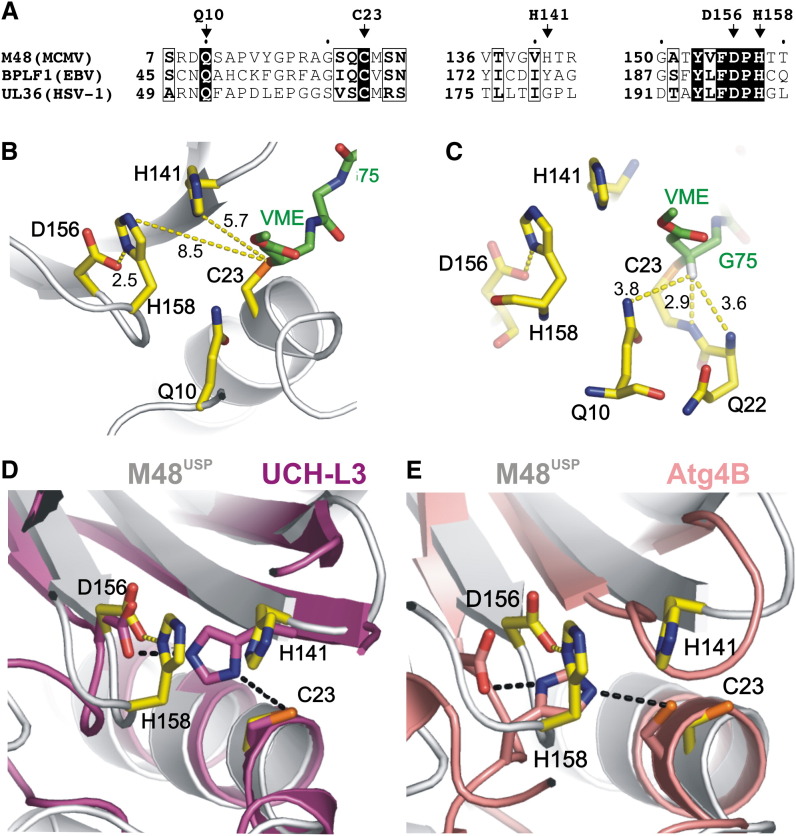
(A) Sequence alignment of herpesvirus-encoded USPs M48 (MCMV), UL36 (HSV-1), and BPLF1 (EBV). Conserved residues are boxed, and conserved catalytic triad residues (C23, D156, and H158) and Q10 of the oxyanion hole residues are indicated and shown in (B) as yellow sticks. Furthermore, H141, which is not conserved but might participate in catalysis, is also shown. The covalently bound UbVME is represented by green sticks. Note that the catalytic triad is misaligned because both histidines are >5 Å from C23.
(C) The proposed oxyanion hole is occupied by the hydrogen atom of the β carbon atom of the former vinylmethylester moiety, thus resembling a catalytic intermediate except that the oxyanion is replaced by a hydrogen. The Q10 side chain and NH groups of C23 and Q22 are <4 Å from this hydrogen atom and are thus likely to coordinate the oxyanion during catalysis.
(D) Superposition of the active sites of M48USP (yellow) and UCH-L3 (pink). H141 corresponds to the location of the UCH-L3 catalytic histidine, and a conformational change of H141 may result in a similar functional catalytic triad alignment as observed for UCH-L3.
(E) Superposition of catalytic triad residues of M48USP (yellow) and human Atg4B (salmon), the only known cysteine protease structure that features a catalytic triad arrangement that aligns its catalytic histidine with M48USP H158. A different rotamer conformation for M48USP H158 may therefore yield an aligned catalytic triad.