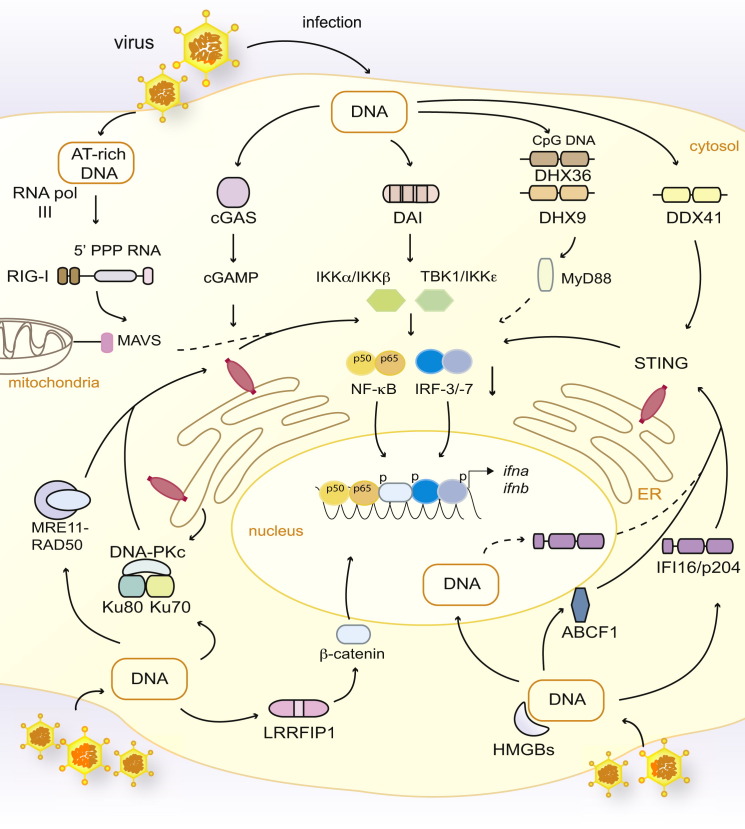
Figure 4.
Putative Intracellular DNA Sensors Involved in IFN-α and IFN-β Induction
DNA present in the cytosol after viral infection induces the production of type I IFNs through a central signaling cascade involving STING, which serves as a scaffold for the phosphorylation of IRF-3 by the kinase TBK1. It was recently demonstrated that the cytosolic nucleotidyltransferase cGAS binds DNA and synthesizes the formation of a cyclic-GMP and cyclic-AMP hybrid termed cGAMP, which directly binds to and activates STING. Cytosolic DNA is reported to engage a number of additional receptors, including DAI, human IFI16 (or mouse p204), and the helicases DDX41, DHX36, and DHX9. DHX36 and DHX9 appear to be specific to CpG DNA and are reported to signal via MyD88. HMGB1, HMGB2, and HMGB3 have also been shown to promote cytosolic DNA responses. Data also suggest that ABCF1 binds DNA and interacts with HMGB2 and p204 to stimulate innate immune responses. Moreover, AT-rich DNA can be transcribed by RNA pol III into 5′-PPP-containing RNA (5′ PPP RNA), which serves as a RIG-I agonist. LRRFIP1 senses cytosolic DNA and phosphorylates β-catenin, which translocates to the nucleus and promotes IFN-β transcription. The DNA-PKc-Ku70-Ku80 and MRE11-RAD50 complexes, involved in DNA-damage responses, have additionally been suggested to bind cytosolic DNA and promote STING-dependent type I IFN responses. Ku70 is further reported to trigger the expression of type III IFNs in an IRF-1- or IRF-7-dependent manner in response to cytosolic DNA (not depicted; Zhang et al., 2011a). Proteins involved in DNA-damage responses, as well as IFI16 and RNA pol III, are abundantly present in the nucleus, highlighting the possibility that DNA sensing might also occur in that organelle (only depicted here for IFI16). Dashed lines indicate indirect or possible signaling, and “p” indicates phosphorylated proteins.