Figure 5.
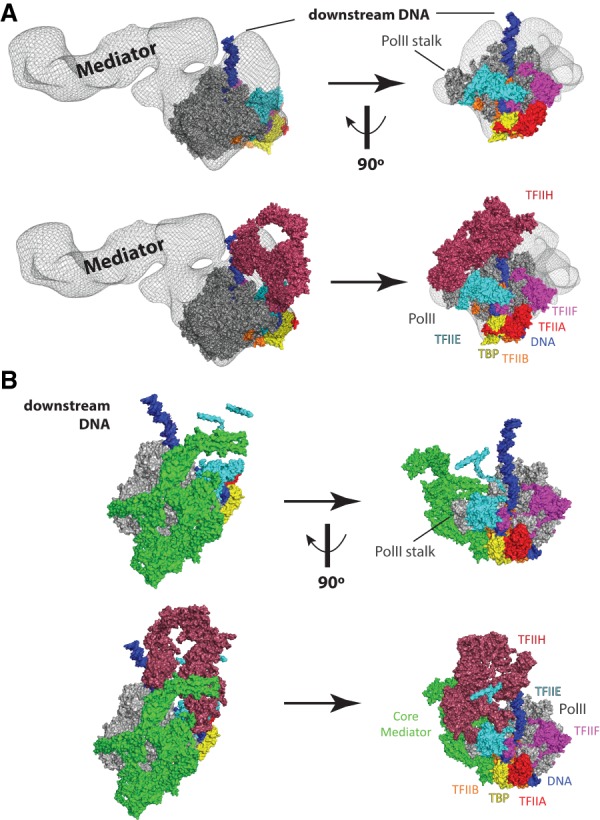
PIC structural models that include the Mediator complex. (A) Model of a partial human PIC that includes Mediator. Figure was prepared by rendering a Mediator–Pol II–TFIIF cryoEM density (Bernecky et al. 2011) in PyMol as a semiopaque black mesh, then visually aligned to a human PIC structure (He et al. 2016). Colors for each PIC factor are identical to Figure 1. The top row shows two views of the complex without TFIIH, and the bottom row shows the same views with TFIIH. Structural remodeling is likely upon binding TFIIH, as clashes are evident in this artificially docked model. The differences between yeast (see B) and human Mediator reflect the much larger size of the human Mediator complex (Table 1). However, the orientation of the human Mediator complex modeled in the human PIC is distinct from the yeast PIC. These differences could result from true differences in PIC structure (yeast vs. human) or could simply result from the fact that the human PIC model is not derived from a single complete structural assembly, as done for the yeast PIC (Schilbach et al. 2017). Adapted from PDB 5IY7 (He et al. 2016) and EMD-5343 (Bernecky et al. 2011). (B) Structure of a yeast PIC (S. cerevisiae), shown in identical orientations with A, based on alignment of Pol II. Here, a core Mediator complex is shown in green, whereas all other PIC factors are shown in the same colors as A. The top row shows two views of the complex without TFIIH, and the bottom row shows the same views with TFIIH. The different orientation of downstream DNA (vs. A) reflects potential structural differences between yeast and human PICs. Note, however, that A is a hypothetical model that merges two different structures, whereas B represents cryoEM data from a single structure (Schilbach et al. 2017). Adapted from PDB 5oqm (Schilbach et al. 2017).
