Abstract
Porcine epidemic diarrhea virus (PEDV), a member of the genus Alphacoronavirus, has caused significant damage to the Asian and American pork industries. Coronavirus 3C-like protease (3CLpro), which is involved in the processing of viral polyproteins for viral replication, is an appealing antiviral drug target. Here, we present the crystal structures of PEDV 3CLpro and a molecular complex between an inactive PEDV 3CLpro variant C144A bound to a peptide substrate. Structural characterization, mutagenesis and biochemical analysis reveal the substrate-binding pockets and the residues that comprise the active site of PEDV 3CLpro. The dimerization of PEDV 3CLpro is similar to that of other Alphacoronavirus 3CLpros but has several differences from that of SARS-CoV 3CLpro from the genus Betacoronavirus. Furthermore, the non-conserved motifs in the pockets cause different cleavage of substrate between PEDV and SARS-CoV 3CLpros, which may provide new insights into the recognition of substrates by 3CLpros in various coronavirus genera.
Keywords: Corona virus, PEDV 3CLpro, Crystal structure, Dimerization, Substrate specificity
Highlights
-
•
The substrate binding mechanism of PEDV 3CLpro has been characterized.
-
•
The large buried surface area is responsible for dimerization of PEDV 3CLpro.
-
•
Non-conserved motifs cause different cleavage between PEDV and SARS-CoV 3CLpros.
1. Introduction
Porcine epidemic diarrhea virus (PEDV), which belongs to the genus Alphacoronavirus in the family Coronaviridae, causes severe diarrhea, vomiting, dehydration and high mortality in neonatal piglets (Song and Park, 2012, Wood, 1977). PED was first observed in Europe in 1971 (Oldham, 1972). PEDV was isolated in Belgium in 1976 (Pensaert and de Bouck, 1978). Since PEDV was first identified, outbreaks have been reported in many swine-producing countries, notably in Europe and Asia. PEDV has recently re-emerged, with outbreaks in Asia and North America resulting in enormous economic losses (Pasick et al., 2014, Vlasova et al., 2014, Wang et al., 2014b). Continuous vaccine efforts have been made and some advances have been achieved since the outbreak (Collin et al., 2015, Song et al., 2015).
There are four genera within Coronaviridae: Alphacoronavirus, Betacoronavirus (A, B, C and D) (de Groot et al., 2012), Gammacoronavirus, and Deltacoronavirus (Adams and Carstens, 2012). Transmissible gastroenteritis virus (TGEV), another Alphacoronavirus, causes severe and often fatal diarrhea in young pigs (Garwes, 1988). The severe acute respiratory syndrome (SARS) coronavirus (lineage B Betacoronavirus) causes a life-threatening disease with a mortality rate of 11% (Weiss and Navas-Martin, 2005). Another novel lineage C Betacoronavirus, the Middle East respiratory syndrome coronavirus (MERS-CoV), has recently emerged in the Middle East and spread to Europe and other areas, causing a SARS-like infection in humans with a high mortality rate of approximately 40% (Chan et al., 2013).
The coronavirus replicase gene consists of two large open reading frames (ORF1a and ORF1b) located at the 5′ end of the genome. ORF1a encodes the polyprotein pp1a, whereas ORF1a and ORF1b together encode pp1ab (Hegyi et al., 2002, Ziebuhr et al., 2000, Ziebuhr, 2005). The coronavirus 3C-like protease (3CLpro; non-structural protein 5, Nsp5) is part of the polyproteins pp1a and pp1ab, and this enzyme is encoded by ORF1a. With few exceptions, coronaviruses encode two papain-like proteases (called PL1pro and PL2pro, respectively), which are responsible for the cleavage of the N-proximal regions of the polyprotein. The central and C-proximal regions of the polyprotein are cleaved by 3CLpro at 11 conserved sites, and the products are essential for viral replication (Thiel et al., 2001, Thiel et al., 2003, Ziebuhr et al., 2000). Thus, 3CLpro is an appealing target for the design of anti-coronavirus therapies.
Coronavirus 3CLpro employs conserved cysteine and histidine residues, which serve as the principal nucleophile and general acid-base catalyst, respectively, at its catalytic site (Anand et al., 2002, Hegyi et al., 2002, Lu et al., 1995, Thiel et al., 2003, Ziebuhr et al., 2000). An asparagine is the third member of the catalytic triad in proteases of the papain family. Chymotrypsin and other members of this serine protease family possess a catalytic triad (Anand et al., 2002). However, in coronavirus 3CLpro, there is no third residue involved in catalysis (Hegyi et al., 2002, Liu and Brown, 1995, Lu and Denison, 1997). Notably, a buried water molecular hydrogen-bonded to three surrounding residues takes the place that is normally occupied by the side chain of the third member of the catalytic triad (Anand et al., 2002, Anand et al., 2003, Yang et al., 2003). Coronavirus 3CLpro recognizes a conserved site containing a hydrophobic residue (preferably L) at the P2 position, a Q at the P1 position, and a small aliphatic amino acid residue (S, G, A) at the P1′ position (Hegyi and Ziebuhr, 2002, Hsu et al., 2005, Ziebuhr et al., 2000). The previously reported nomenclature system was used to describe the residue sites of the peptide substrates and the substrate binding sites of the protease (Schechter and Berger, 1967).
Dimerization is a commonly used strategy for regulating viral protease activity. The formation of a dimer can be the mechanism for enzyme activation. Conversely, dimerization can inhibit an active monomeric enzyme (Marianayagam et al., 2004). In terms of coronavirus 3CLpros, only the dimeric form is functional (Chen et al., 2008, Li et al., 2010, Shi et al., 2008). In the structure of TGEV 3CLpro, the N-terminus of one monomer helps shape the S1 pocket and the oxyanion hole of the opposite monomer; thus, dimerization is essential for its catalytic activity (Anand et al., 2002). The dimerization and trans-cleavage activity of SARS-CoV 3CLpro are completely inhibited by the E290R and R298E (located at the C-terminal helix) variants and partly inhibited by the R4E (located at the N-terminal loop) variant (Hilgenfeld, 2014).
To investigate the properties of PEDV 3CLpro, we determined the crystal structure of PEDV 3CLpro, as well as that of an inactive PEDV 3CLpro variant (C144A) bound to a peptide substrate. In addition, we performed biochemical analyses and structural comparisons to provide further insights into the dimerization and substrate specificity of coronavirus 3CLpro.
2. Results and discussion
2.1. Overall structure of PEDV 3CLpro
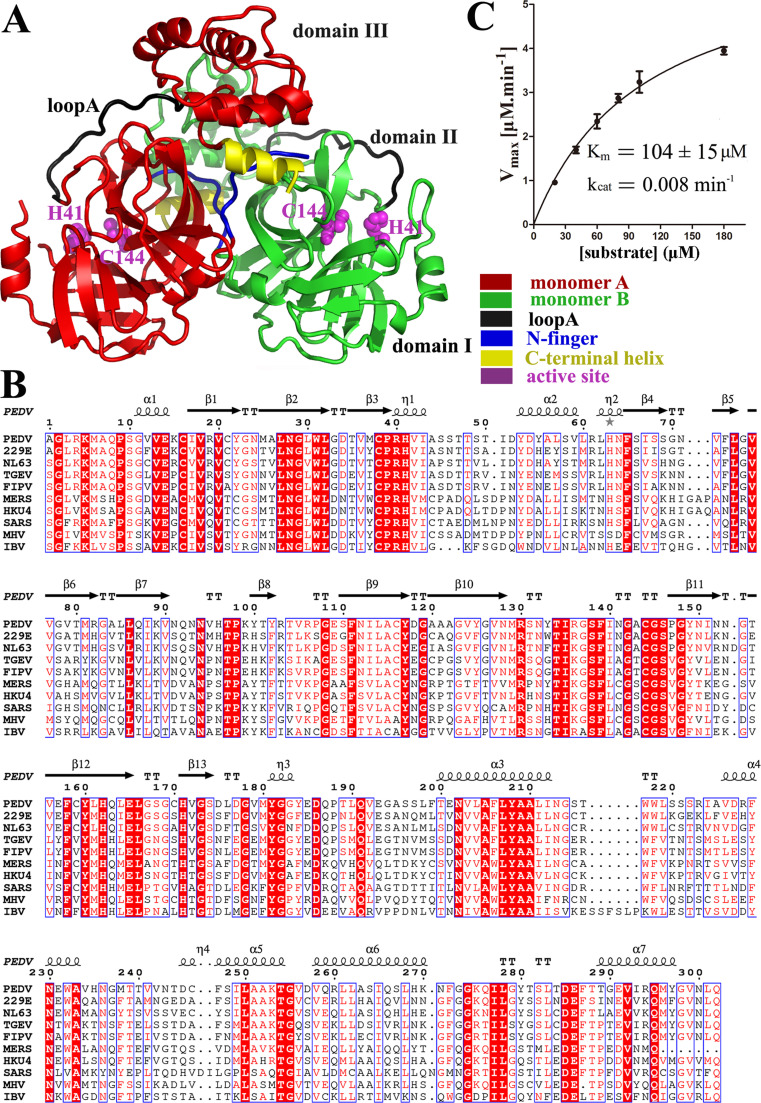
The PEDV 3CLpro crystal structure was determined at a high resolution (1.65 Å) in the space group, P21 ( Fig. 1). Two molecules are observed in each asymmetric unit, with a root mean square deviation (RMSD) of 0.24 Å for 186 superimposed Cα atoms of domain I and domain II, 0.89 Å for 97 superimposed Cα atoms of domain III (computed through the PDBeFold service on the European Bioinformatics Institute website); the two protomers are oriented at approximately right angles to each other. Each monomer features three domains: domain I (residues 1–97), domain II (residues 98–186), and domain III (residues 202–298). Each of the first two domains exhibits an antiparallel β-barrel structure, which is similar to the 3CLpros from other coronaviruses (Anand et al., 2002, Needle et al., 2015, Yang et al., 2003). Domain III consists of five α helices and is connected to domain II by a long loop (residues 187–197, loop A in Fig. 1(A)). As observed in all of the reported 3CLpro structures, the substrate-binding site near the catalytic dyad is located in a cleft between domains I and II.
Fig. 1.
Crystal structure and sequence alignment of PEDV 3CLpro. A. The overall structure of the PEDV 3CLpro homodimer. Loop A, the N-finger, the C-terminal helix and the active site are shown in black, blue, yellow and magenta, respectively. Loop A, which connects domain II to domain III, is labeled. Catalytic residues H41 and C144 are represented as spheres. B. The structure-based sequence alignment of several coronavirus 3CLpros. The sequence alignment was conducted with Clustal W and the figure was prepared with ESPript 3 (Robert and Gouet, 2014). Residues conserved in all 3CLpros are shown in white on a red background. Residues conserved in most of the sequences are shown in red and boxed with a white background. C. Kinetic parameters of PEDV 3CLpro. The fluorogenic substrate (Dabcyl-YNSTLQ↓AGLRKM-E-Edans) was used in the assays. The initial rates of the protease under different substrate concentrations were used to calculate the kinetic parameters by fitting with the Michaelis-Menten equation using GraphPad Prism5.
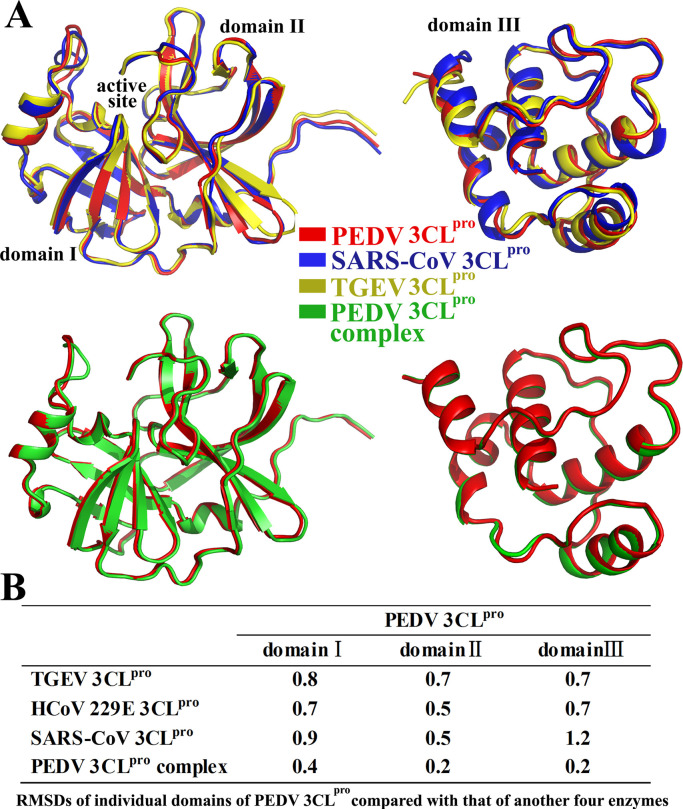
Structural alignment of the domains of several coronavirus 3CLpros gives an overall view of the comparison ( Fig. 2). The structures show high similarity, especially in the area around the active site, and several small shifts are found mainly in domain III (Fig. 2(A)). The RMSD value (1.2 Å) of domain III between SARS-CoV 3CLpro and PEDV 3CLpro is higher than that (0.7 Å) between the two Alphacoronavirus 3CLpros and PEDV 3CLpro. The RMSD values of the first two domains between different coronavirus 3CLpros are relative small (all around 0.7 Å) (Fig. 2(B)). These results reveal that SARS-CoV 3 CLpro (Betacoronavirus) demonstrates a relatively high number of structural differences compared with the Alphacoronavirus 3CLpros, especially in domain III; the first two domains (chymotrypsin-like cores) show higher degrees of conservation compared with that of domain III among different genera of coronavirus.
Fig. 2.
Structure alignment of individual domains of several coronavirus 3CLpros. A. Schematic overview of superimposed domains of one monomer from several coronavirus 3CLpros (PEDV 3CLpro, PDB Entry: 4XFQ; SARS-CoV 3CLpro, PDB Entry: 2H2Z; TGEV 3CLpro, PDB Entry: 1LVO; PEDV 3CLpro complex, PDB Entry: 4ZUH; HCoV 229E 3CLpro, PDB Entry; 1P9S). B. RMSD values of individual domains of the PEDV 3CLpro compared with that of four additional enzymes (computed using the PDBeFold service on the European Bioinformatics Institute website).
The imidazole ring of H41 of the catalytic dyad adopts a different orientation from that observed in most current coronavirus 3CLpro structures. The imidazole ring of H41 is not in contact with the corresponding nucleophile C144 in PEDV 3CLpro (Fig. S1). However, this configuration may be an artifact caused by the low pH (4.2) of crystallization. At this pH, the histidine residue may be protonated and become unable to interact with the sulfhydryl of C144. Such a pH-effect has been previously described for the 3C protease of enterovirus 68 (Tan et al., 2013).
2.2. The dimerization of PEDV 3CLpro
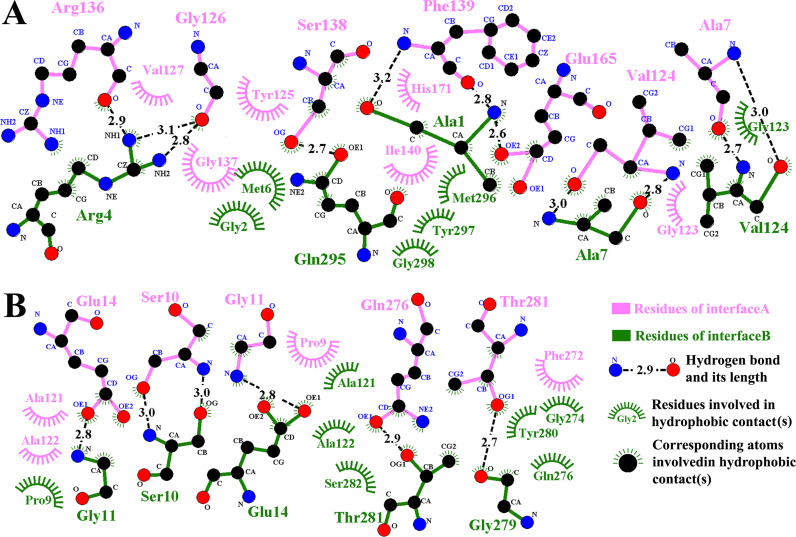
Tablesimeric form has consistently been shown to be the catalytically active form for all coronavirus 3CLpros studied to date. At the dimer interface of PEDV 3CLpro, the N-terminal finger (8 residues of the N-terminus) assembles into the dimerization surface, interacting with domain II of the opposite monomer and the C-terminal helix of the same monomer (Fig. 1(A)). A total surface area of 2085 Å2 is buried upon dimerization (computed using PDBePISA tool, http://pdbe.org/pisa/). In this molecule, 16 residues from monomer A are found to interact (hydrogen bonds or salt bridges) with 16 residues from monomer B at the dimer interface ( Table 2 and Fig. 3). All the residues are conserved among the three Alphacoronavirus 3CLpros, except three substitutions for the A1, G279 and T281 residues (Table 2). Meanwhile, extensively hydrophobic interactions appear at the dimer interface. The interactions are mainly observed between the N-terminal residues of one monomer and domain II and domain III of the opposite monomer. The intraprotomer interactions between the two N-terminal loops and between the two domain III are also involved in the dimerization (Fig. 3(B)).
Table 2.
The comparison of residue-residue interactions at the dimer interface from PEDV 3CLpro with these from other 3CLpros.
|
PEDV 3CLpro |
TGEV 3CLpro |
HCoV 229E 3CLpro |
SARS-CoV 3CLpro |
||||
|---|---|---|---|---|---|---|---|
| Monomer A | Monomer B | Monomer A | Monomer B | Monomer A | Monomer B | Monomer A | Monomer B |
| 1A | 139F,165E | 1S | 139F, 163E | 1A | 139F,165E | 1S | 166E |
| 2G | 138S | 2G | 138S | 2G | 138S | 2G | 139S, 140F |
| 4R | 126G, 286E | 4R | 126G, 286E | 4R | 126G, 286E | 4R | 127Q, 139S, 290E |
| 7A | 124V | 7A | 124V | 7A | 124V | 7A | 125V |
| 10S | 10S | 10S | 10S | 10S | 10S, 14E | 10S | 10S |
| 11G | 14E | 11G | 14E | 11G | 14E | 11G | 14E |
| 14E | 11G | 14E | 11G | 14E | 10S, 11G | 14E | 11G |
| 124V | 7A | 124V | 7A | 124V | 7A | 125V | 7A |
| 126G, 286E | 4R | 126G, 286E | 4R | 126G, 286E | 4R | 127Q, 290E | 4R |
| 138S | 2G,295Q | 138S | 2G, 295Q | 138S | 2G,295Q | 139S | 2G, 299Q |
| 139F, 165E | 1A | 139F, 165E | 1S | 139F, 165E | 1A | 166E | 1S |
| 279G | 281T | 279S | 281G | 281S | 282S | ||
| 281T | 276Q | 281G | 279S | 282S | 281S | ||
| 295Q | 138S | 295Q | 138S | 295Q | 138S | 299Q | 139S |
| 298R | 123S | ||||||
The PDB identifiers for the four structures are 4XFQ, 1LVO, 2ZU2 and 1UK4. The hydrogen-bond and hydrophobic interactions are all analyzed in PyMOL (Schrödinger, 2006). For the three Alphacoronavirus 3CLpros, the residue-residue interactions are conserved except the two interactions (in purple bold) between domain III of each monomer. However, these two interactions are not observed in SARS-CoV 3CLpro, and the interaction between 298R and 123S is not found in the Alphacoronavirus 3CLpros.
Fig. 3.
A structural diagram of the distribution of hydrophobic and hydrophilic interactions at the dimer interface. A. The figure was generated using the LigPlot+ program. The residues of interface A and interface B are colored in pink and olive green, respectively. The carbon, nitrogen, and oxygen atoms are shown as black, blue and red circles, respectively. Hydrogen bonds are shown as black dashed lines labeled with the distance between the donor and corresponding acceptor atom. Hydrophobic interactions are demonstrated by arcs with spokes radiating toward the atoms (with spokes around) or residues (shown as arcs with spokes) they contact. B. Another interacting patch at the interface that is important for dimerization.
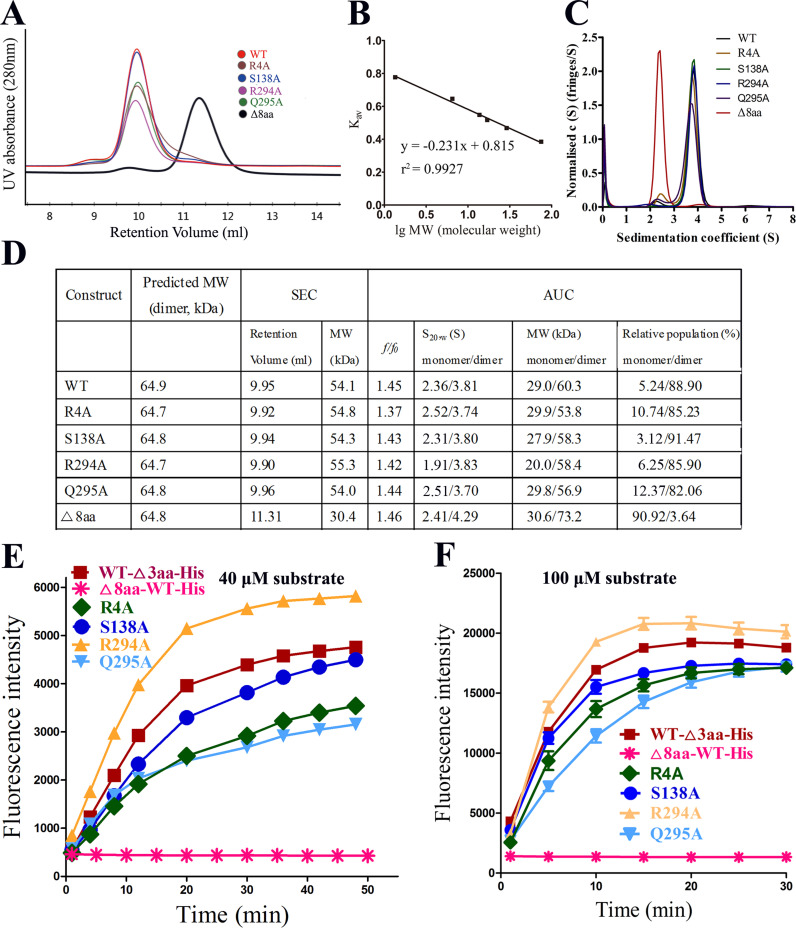
To explore the relationship between oligomerization and enzymatic activity, four variants (R4A, S138A, R294A, and Q295A) and an N-finger deletion variant at the PEDV 3CLpro dimer interface were constructed. The elution peaks of the four variants show similar retention volumes to that of the wild type (WT) at 9.95 ml (approximately 54 kDa), which represents a dimer form similar to that of WT PEDV 3CLpro ( Fig. 4(A), (B) and (D)). However, the elution peak of the N-finger deletion variant (Δ8aa) shows a retention volume of 11.31 ml (approximately 30.4 kDa), which represents the monomer. The oligomerization of the proteins was further confirmed through sedimentation velocity experiments (Fig. 4(C) and (D)). The results were consistent with that of the gel filtration chromatography analysis. The majority (88.90%) of wild type PEDV 3CLpro exists as dimers, and few (5.24%) exists as monomers. The four variants still primarily exist as dimers, however the relative populations of the variants monomers, except S138A, increased slightly. The majority (90.92%) of the N-finger deletion variant exists as monomers. These data indicate that wild type PEDV 3CLpro primarily exists as dimers in solution and the engineered variants (R4A, S138A, R294A, and Q295A) show little effect on the monomer-dimer equilibrium. The N-finger deletion absolutely disrupts the dimerization.
Fig. 4.
Mutational studies of several residues at the dimer interface. A. Size exclusion chromatography (SEC) analyses of the four site variants and the N-terminal-deletion variant (Δ8aa). WT, Δ8aa, Q295A, R294A, S138A and R4A are shown in red, black, orange, blue, yellow and green, respectively. B. Calibration curve. Conalbumin, 75 kDa; carbonic anhydrase, 29 kDa; equine myoglobin, 17 kDa; ribonuclease A, 13.7 kDa; aprotinin, 6.5 kDa; vitamin B12, 1.35 kDa (Bio-Rad and GE Healthcare) were used to calibrate the column. The values of the Y-axis were calculated using the equation Kav=(Ve−V0)/(Vt−V0). C. Sedimentation velocity analysis of the WT and variants. The major peak for each sample represents the major state of the protease. D. Calculated molecular masses based on the results of SEC and AUC assays, respectively. Δ8aa primarily exists as monomers. E. FRET-based mutational studies of the residues that may be involved in the dimerization of PEDV 3CLpro. The S138A, R4A, and Q295A variants exhibited reduced catalytic activity to various degrees (1, 0.74, 0.94, 1.22, 0.66, and 0.09, respectively; the activity of wild type PEDV 3CLpro was taken as 1). None of the three variants caused a great or complete loss of activity, and the catalytic activity of the R294A variant exhibited no reduction. Only the N-finger deletion variant exhibited a complete loss of activity. F. Same as that in panel E but with different substrate concentration.
FRET (Matayoshi et al., 1990) results demonstrate that the mutations reduced the catalytic activity to varying degrees (Fig. 4(E) and (F)). Only the N-finger deletion variant exhibited a complete loss of activity. The large buried surface area is responsible for dimer association of PEDV 3CLpro and one residue substitution may not completely destroy the dimerization. The N-finger residues of coronavirus 3CLpro are key components of the buried interface and are essential for dimerization. Thus, the N-finger deletion mutation destroyed the dimer form and inactivated PEDV 3CLpro.
In SARS-CoV 3CLpro, a hydrophobic interaction between the side chains of M6 and Y126 of the opposite promoter greatly contributes to the stabilization of the dimer conformation (Wei et al., 2006). At the same time, the backbone oxygen of M6 and the nitrogen atom of the side chain NH2 of R298 in the same monomer form a hydrogen bond (2.94 Å), which can be further connected to the catalytic machinery of the opposite protomer through a two-step relay (Shi et al., 2008). Thus, R298 is very important for the catalytic activity of SARS-CoV 3CLpro. As a result, the R298A mutation in the SARS-CoV 3CLpro caused a dimer-monomer switch and inactivated the enzyme (Shi et al., 2008). However, the homologous variant (R294A) in PEDV 3CLpro remained a dimer and an active enzyme (Fig. 4(E) and (F)). In the PEDV 3CLpro structure, R294 shows no interactions with any residues from another monomer. The result shows that R294 is not a key component involved in the dimerization of PEDV 3CLpro.
Despite high sequence and structure similarities, differences in dimerization exist among 3CLpros from different coronavirus genera. As in MERS-CoV 3CLpro, non-conserved residues far from the dimer interface might be involved in dimer formation of a weakly associated dimer (Tomar et al., 2015). These structural differences may lead to differences in the dimerization and enzymatic properties of 3CLpros from different coronavirus groups.
2.3. Overall structure of the PEDV 3CLpro C144A variant in complex with the peptide substrate
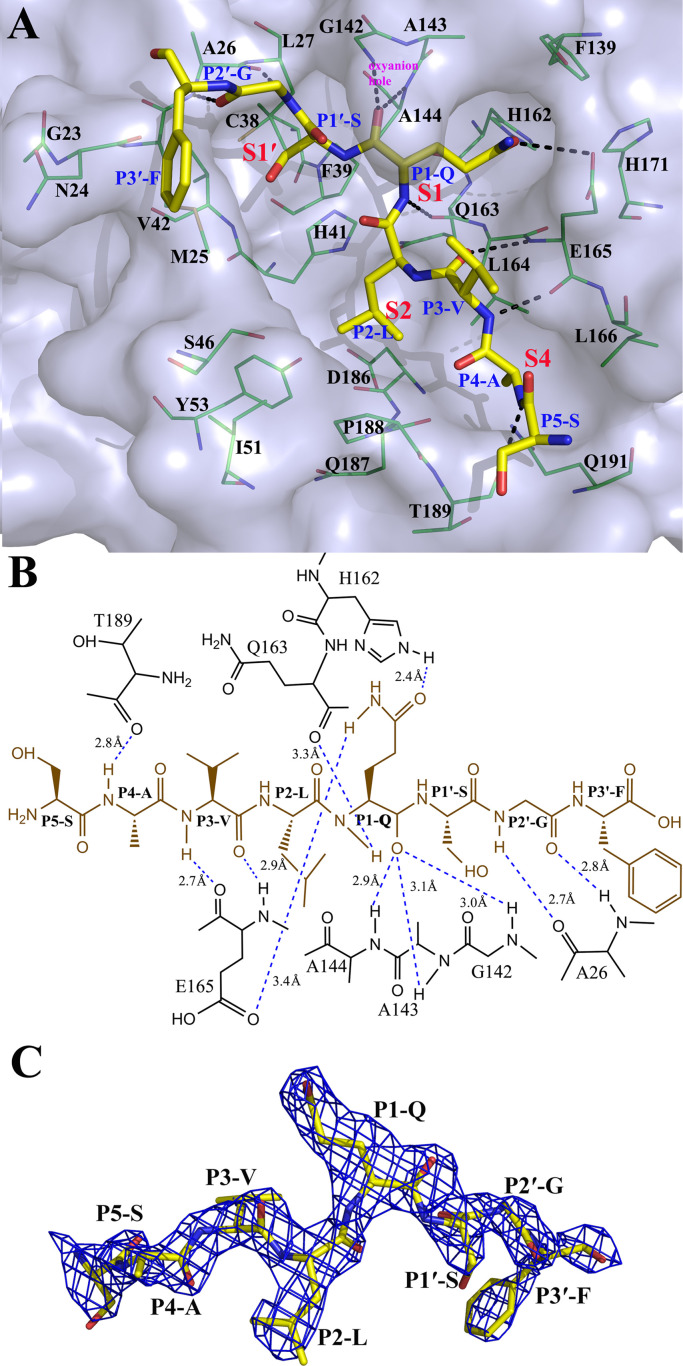
In order to investigate further the substrate recognition mechanism of PEDV 3CLpro, we synthesized a peptide (TSAVLQ↓SGFRK) (Nanjing GenScript Company) for co-crystallization and determined the crystal structure of the C144A variant complex at 2.2 Å resolution in the space group, P21. Two 3CLpro molecules are observed in the asymmetric unit, and the two monomers have the similar overall structures (RMSD of 0.25 Å for 186 superimposed Cα atoms of domain I and domain II, 0.92 Å for 99 superimposed Cα atoms of domain III). However, the first two domains show low RMSD values, it seems that they are more stable than the C-terminal helix domain. An eight amino acid peptide (SAVLQ↓SGF) is observed in only one monomer according to the density map ( Fig. 5), but the substrate-binding sites of the two monomers are almost identical. The first two domains show almost no differences and domain III shows slight structure deviation. However, the substrate lies in the cleft between the first two domains. Thus, the absence of the peptide in another monomer may be caused by the poor density map or the crystal package. The four residues P4–P1 form an antiparallel β sheet with residues 162–166, and residues P5–P4 form an antiparallel β sheet on the other side with residues 188–190 of the long loop A, that links domain II and III (Fig. 5(A)). The P1′ to P3′ strand extends into the solvent, and P2′–G forms a short antiparallel strand interaction with A26.
Fig. 5.
Structure of a PEDV 3CLpro variant (C144A) bound to a peptide substrate. A. Three-dimensional structure of the substrate-binding pockets with a peptide. The residues of the pockets are represented in green as a stick diagram, and the substrate is shown in yellow as a stick diagram. The S1, S2, S4 and S1′ pockets are labeled. The residues are all labeled. The nitrogen atoms are shown in blue, and oxygen atoms are shown in red. Hydrogen bond interactions are shown as black dashed lines. B. Diagram of the detailed molecular interactions between the substrate and the protease. The peptide substrate is shown in brown. Hydrogen bonds are shown as blue dashed lines, and the hydrogens were connected to their acceptors by the dashed lines. The distances labeled reflect the distances between the donor and the acceptor for all hydrogen bonds. Ceratina electron density map of the peptide substrate (2Fo-Fc, contoured at 1.0 σ).
2.4. Details of the substrate-binding sites of PEDV 3CLpro
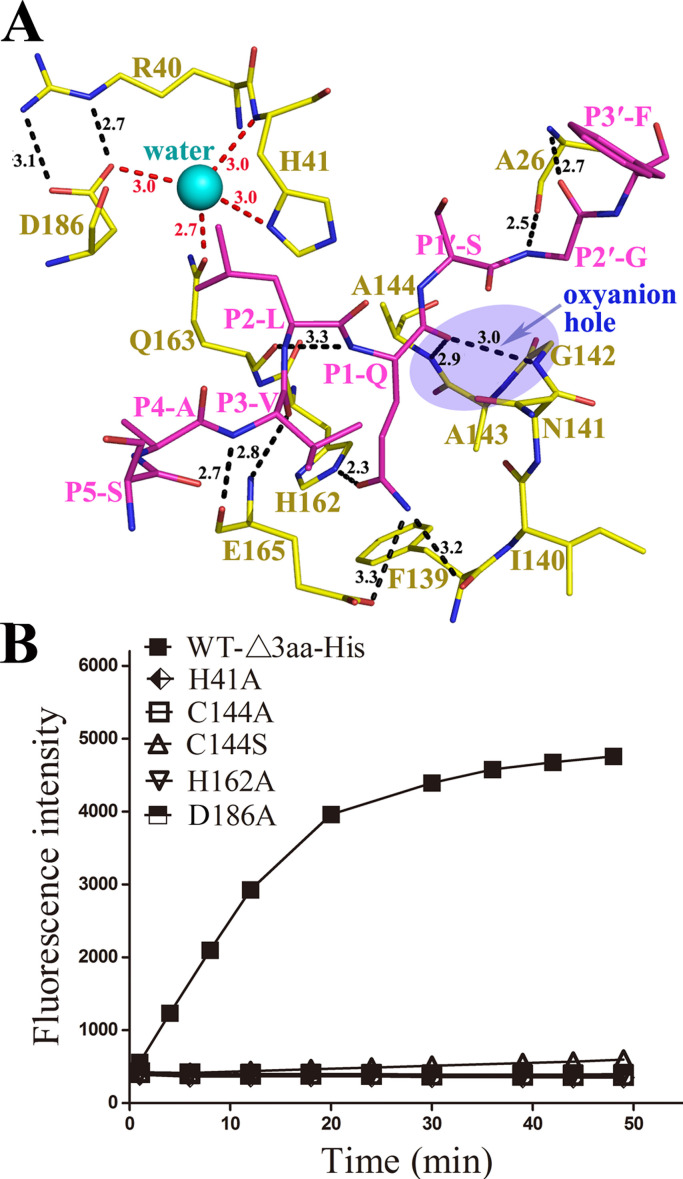
Coronavirus 3CLpro recognizes a conserved residue (glutamine) at the P1 position for efficient hydrolysis (Hegyi et al., 2002, Ziebuhr and Siddell, 1999, Ziebuhr et al., 2000). The side chain of the conserved P1-Q fits comfortably in the pocket (Fig. 5(A)), stabilized by two hydrogen bonds: one between the Nε2 atom of H162 and the Oε1 atom of P1-Q and another between the side chain oxygen of E165 and the Nε2 atom of P1-Q (Fig. 5(B)). The main chain amides of G142 and A144 form an oxyanion hole that stabilizes the carbonyl oxygen of P1-Q. The main chain nitrogen atom of P1-Q forms a hydrogen bond with the main chain oxygen of Q163. The S2 sub-site of PEDV 3CLpro consists of P188, L164, I51, S46 and H41, similar to that of TGEV 3CLpro, which is lined with the side chains of P188, L164, I51, T47 and H41 (Anand et al., 2002). The side chains of these residues form a hydrophobic pocket that can accommodate a P2 residue with a large side chain, such as leucine or methionine (Fig. 5(A)). The main chain of P3-V forms two hydrogen bonds with the main chain of E165, and the side chain of P3-V is exposed to the solvent (Fig. 5(A)). Since the side chain of P3 Val is pointing to the surface and therefore to the solvent, the charged character of the P3 residue has no difference or preference. The P4 pocket consists of a series of hydrophobic amino acids and may prefer residues with high hydrophobicity. A hydrogen bond is found between the main chain NH of P4-A and the carbonyl oxygen of T189. The PEDV 3CLpro S1′ sub-site is composed of M25, A26, L27, C38, P39, H41 and V42. Small residues such as serine, glycine and alanine are preferred in the small S1′ pocket. The main chain of P2′-G forms two hydrogen bonds with the main chain atoms of A26 (Fig. 5(B)). Due to the poor density of P3′-F, its carbonyl group and the side chain phenyl group could easily be interchanged during structure refinement. The conservation in the substrate binding pockets provides a basic model for the design of potential antiviral agents (Yang et al., 2005). In the PEDV 3CLpro structure, the imidazole ring of H41 forms an abnormal conformation due to the low crystallization pH. However, in the complex structure of the active-site variant (C144A), the H41 imidazole ring is recovered (Fig. S1). The entire imidazole ring lies parallel with the side chain of P2-L and interacts through hydrophobic effects, and the Nε2 of 41 h becomes closer (3.3 Å) to the peptide bond between P1-Gln and P1′-Ser.
Part of the detailed interaction network is shown in Fig. 6(A). A water molecule is hydrogen-bonded to three residues (H41, Q163 and D186). Notably, the buried water molecule takes the place that is normally occupied by the side chain of D102 in the catalytic triad of the chymotrypsin and other members of this serine protease family. The oxyanion hole that stabilizes the main chain oxygen of P1-Q is made up of the main chain amides of G142 and A144. To determine whether the residues around the catalytic dyad are essential for hydrolytic activity, we performed proteolytic activity analyses of the variants (H41A, C144A, C144S, H162A and D186A). The results demonstrate that the fluorescence values of WT PEDV 3CLpro increase in a time-dependent manner, whereas none of the variants exhibit enzymatic activity (Fig. 6(B)). This finding indicates that the catalytic dyad of PEDV 3CLpro is essential for its proteolytic activity. Additionally, H162 and D186 in the substrate binding site are both essential, which are conserved among coronavirus 3CLpros (Fig. 1(B)).
Fig. 6.
Key residues for enzyme catalytic activity. A. A structural diagram of several key residues involved in enzyme catalytic activity. The residues of the protease are colored yellow, and the substrate is colored magenta. The residues of the substrate are labeled and shown as stick diagrams. Hydrogen bond interactions are shown as dashed lines, along with the bond distance. A water molecule buried by three residues (H41, Q163 and D186) is shown as a blue sphere. The oxyanion hole region is shown as a subtransparent blue oval. B. Fluorescence profiles of the hydrolysis of the fluorogenic substrate by WT-Δ3aa-His and the variants of PEDV 3CLpro. The enhanced fluorescence intensity was monitored for 60 min. All five site-directed variants showed nearly no activity compared with WT-Δ3aa-His.
A His-tag was added to the C-terminus during construction. Unfortunately, we could not see the spatial orientation of the tag due to the poor density map. The WT-∆3aa variant without a His-tag was also constructed, and the protease performed similar enzymatic activity to that of the WT-∆3aa-His variant (data not shown). Therefore, the tag may not cause major effect on both the activity and on the conformation of the enzyme.
2.5. None-conserved motifs cause differences in substrate specificity
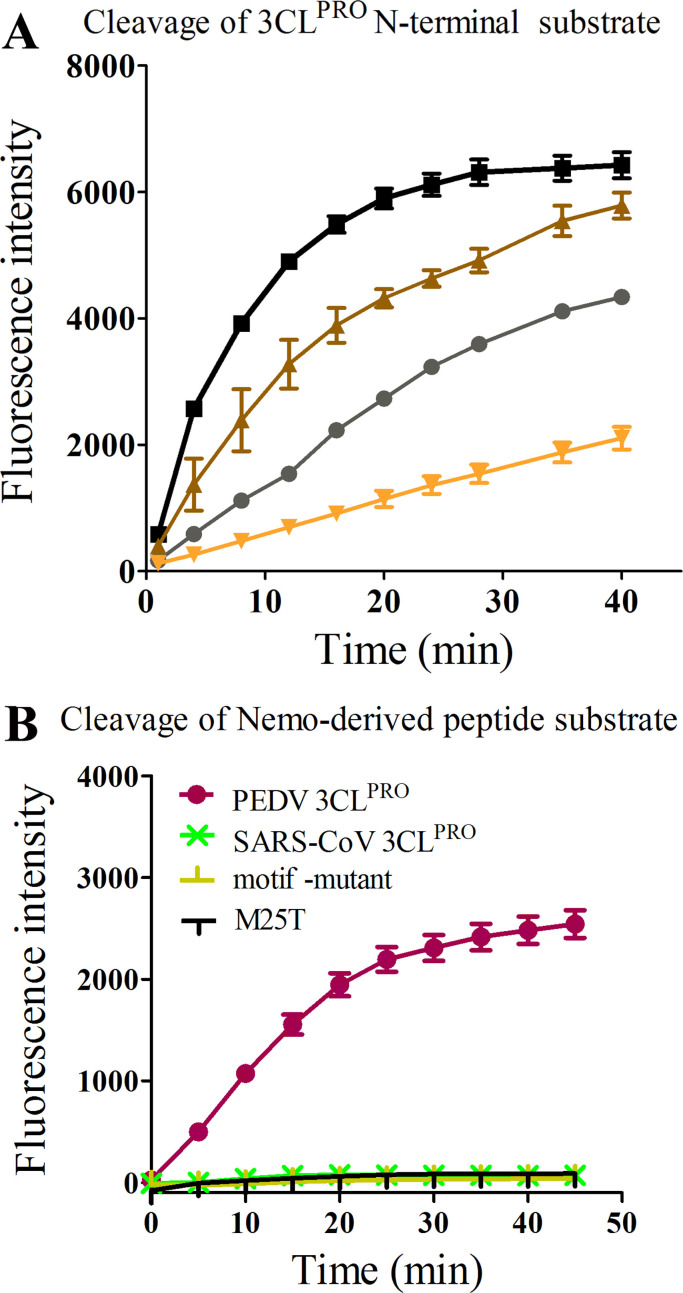
In general, Coronavirus 3CLpros recognize a conserved cleavage site. To determine whether the two proteases from the first two coronavirus genera have differences in substrate specificities, two fluorogenic peptide substrates (Dabcyl-KTSAVLQ↓SGFRKME-Edans and Dabcyl-YNSTLQ↓AGLRKME-Edans contain the N-terminal auto-cleavage sites of PEDV and SARS-CoV 3CLpros, respectively) were introduced in the FRET assays. In addition to processing viral polyproteins, coronavirus 3CLpro has the potential to cleave host proteins during virus-host interactions. PEDV 3CLpro regulates its interferon antagonism by cleaving nuclear transcription factor-kappaB (NF-κB) essential modulator (NEMO) (Wang et al., 2016). The third fluorogenic peptide substrate (Dabcyl-KLAQLQ↓VAYHQE-Edans), which contains the cleavage site derived from NEMO was also applied in the cleavage assays. Both the proteases are able to cleave the two 3CLpro substrates and prefer the SARS-CoV 3CLpro-derived substrate; however PEDV 3CLpro demonstrates better hydrolysis efficiency than SARS-CoV 3CLpro ( Fig. 7(A)). Surprisingly, SARS-CoV 3CLpro is unable to cleave the NEMO-derived substrate, whereas PEDV 3CLpro shows efficient hydrolysis (Fig. 7(B)). Similar results were obtained when the concentration of the proteases increased by 10-fold (data not shown).
Fig. 7.
Peptide substrate cleavage assays. A. Cleavage of coronavirus 3CLpro N-terminal substrate. Two fluorogenic peptide substrates (Dabcyl-KTSAVLQ↓SGFRKME-Edans and Dabcyl-YNSTLQ↓AGLRKME-Edans contain the N-terminal auto-cleavage site of PEDV and SARS-CoV 3CLpro, respectively) were introduced in the FRET assays. (□: SARS-CoV 3CLpro-derived substrate cleaved by PEDV 3CLpro; ○: PEDV 3CLpro-derived substrate cleaved by PEDV 3CLpro; Δ: SARS-CoV 3CLpro-derived substrate cleaved by SARS-CoV 3CLpro; ∇: PEDV 3CLpro-derived substrate cleaved by SARS-CoV 3CLpro). B. Cleavage of NEMO-derived substrate. Similar results were obtained when the concentration of the proteases increased by 10-folds (data not shown). Four proteases used in each assay were labeled in the figure. Motif-mutant and M25T represent two PEDV 3CLpro variants described in the text. Same concentration of the NEMO-derived substrate was used for the four proteases.
To understand the inability of SARS-CoV 3CLpro to cleave the NEMO-derived substrate, we analyzed its substrate preference at each sub-site according to the previous study. Each residue of “KLAQLQ↓VAYHQE” is preferred at the corresponding sub-sites of SARS-CoV 3CLpro, with the exception of the P1′-V, which prefers small residues with side chain volumes of less than 50 Å3 (Ser; Ala; Cys) at the S1′ sub-site (Chuck et al., 2010, Chuck et al., 2011). The PEDV 3CLpro S1′ sub-site is composed of M25, A26, L27, C38, P39, H41 and V42, and those of SARS-CoV 3CLpro are T25, T26, L27, C38, P39, H41 and V42. The hydrophobic side chain of M25 may form hydrophobic interactions with the side chain of P1′-V, which may lead to the tolerance of P1′-V of PEDV 3CLpro. To verify the hypothesis, we replaced the whole motif with that of SARS-CoV 3CLpro (“RVCYGNMA”19–26“QVTCGTTT” ) and also performed the point mutation M25T. It was confirmed that both the variants performed obvious cleavage of the SARS-CoV 3CLpro-derived substrate (data not shown); however, they failed to cleave the NEMO-derived substrate (Fig. 7(B)). It indicates that the motif (19–26), especially the M25 residue, is important for the cleavage of the NEMO-derived substrate by PEDV 3CLpro. And PEDV 3CLpro appears to bear residues with larger side chains at the S1′ sub-site than SARS-CoV 3CLpro does.
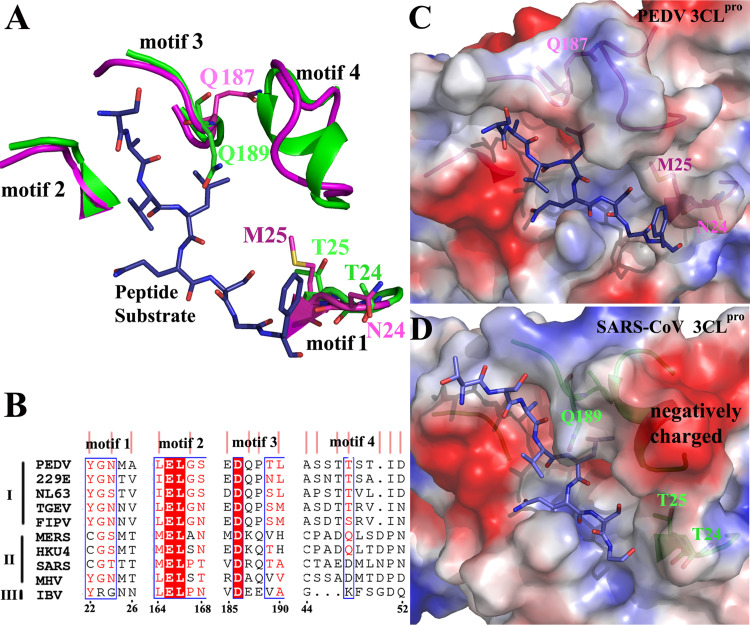
The conserved residues of the sub-sites result in relatively conserved substrate specificity of coronavirus 3CLpros. However, the non-conserved motifs of the pockets may also influence the recognition of the substrate. Except for the motif 1 (residues 22–26) mentioned above, three more motifs are found partly different between PEDV and SARS-CoV 3CLpros (PDB identifier 2Q6G) ( Fig. 8). In motif 1, the two residues (N24 and M25) of PEDV 3CLpro carry different side chains than those (T24 and T25) of SARS-CoV 3CLpro, which result in differences in the pockets mouth (Fig. 8(C) and (D)). Motif 2 of the two proteases consists of partly different residues. In motif 3, the aspartic acid side chains exhibit opposite orientations, which also result in differences in the pocket mouths. The residues of motif 4 obviously differ between the two proteases; they exhibit nearly no sequence homology (Fig. 8(B)), and the SARS-CoV 3CLpro motif 4 contains two negatively charged residues (E47 and D48) (Fig. 8(B) and (C)). We further analyzed the four motifs sequences between Alphacoronavirus and Betacoronavirus 3CLpros (Fig. 8(B)). The four motifs are relatively well conserved in Alphacoronavirus 3CLpros and differ from those of Betacoronavirus. These differences may also lead to the different sizes or depths of the pockets, thus causing the differences in recognition of the substrates.
Fig. 8.
Comparison of the potential substrate-binding motifs of the PEDV and SARS-CoV 3CLpros. A. A cartoon view of the four motifs around the substrate. The PEDV 3CLpro complex structure (magenta) is superimposed with the complex structure of the SARS-CoV 3CLpro variant (C144A) (PDB identifier 2Q6G) (green). The peptide substrate is colored blue. The four motifs and some residues shown as stick diagrams are labeled. B. Sequence alignment of the four motifs of 3CLpros of the three coronavirus groups. The residues that are relatively conserved in Alphacoronavirus 3CLpro but differ from those of Betacoronavirus 3CLpro are marked with pink vertical lines. C. Electrostatic surface potential of the PEDV 3CLpro complex structure. The four motifs are shown as a magenta cartoon and the substrate is shown as blue sticks. Three residues shown as stick diagrams are labeled. D. Electrostatic surface potential of the SARS-CoV 3CLpro complex structure. The four motifs are shown as a green cartoon and the substrate is shown as blue sticks. Three residues shown as stick diagrams are labeled, and the negatively charged area (red) of motif4 is labeled.
3. Conclusion
In summary, we have determined the crystal structure of PEDV 3CLpro alone and in complex with a peptide substrate that contains the N-terminal cleavage site. The strategy for mimicking the peptide substrate side chains to accommodate the corresponding sub-sites can be utilized to design inhibitors (Yang et al., 2005). Thus, our structures may provide the basis for the design of inhibitors against PEDV 3CLpro. The biochemical and structural analyses revealed that PEDV 3CLpro forms a tight dimer, and no variants caused a dimer-to-monomer switch except the N-finger deletion variant. The interaction networks involved in dimer formation of 3CLpro are relatively well conserved among Alphacoronavirus but somewhat different from those of SARS-CoV 3CLpro in Betacoronavirus. Finally, we find that non-conserved motifs in the pockets cause different cleavage of substrate between the PEDV and SARS-CoV 3CLpros; we further analyzed the differences of the motifs between 3CLpros from Alphacoronavirus and Betacoronavirus.
4. Materials and methods
4.1. Cloning, protein expression and purification
The PEDV nsp5 (encoding 3CLpro) gene was cloned from the viral cDNA of the FJZZ strain (GenBank: KC140102.1) via PCR. The forward and reverse primers contained NdeI and XhoI restriction sites, respectively. The PCR product contained a C-terminal His-tag for further protein purification. The PCR product was cloned into the pET42b expression vector. Multiple truncation recombinant plasmids and variants were also constructed. NcoI and XhoI restriction sites were used for the pET28a vector. BamHI and XhoI restriction sites were used for the pET28a-sumo vector. The recombinant WT-Δ3aa-His (the last three amino acids, NLQ, were deleted, and a His-tag was added to the C-terminus) plasmid was used as a template for the following variants: M25T, H41A, H162A, C144A, C144S, D186A and the motif variant. The recombinant His-sumo-WT (a His-sumo fusion tag was added to the N-terminus) plasmid was used as a template for the following site mutations: R4A, S138A, R294A and Q295A. Then, overlap-extension PCR was performed (Ho et al., 1989). All of the recombinant expression plasmids were sequenced, and no unexpected mutations occurred.
For protein expression, the recombinant plasmids were transformed into E. coli BL21 (DE3) and then cultured at 37 °C in LB medium until the OD600 reached 0.6–0.8. Then, 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce protein expression. The cells were harvested after incubation at 27 °C for 7 h, resuspended in PBS and disrupted (ATS AH-1500). The supernatant was filtered and loaded onto a His Trap™ HP column (GE Healthcare), and the C-terminal His-tagged protein was finally eluted using a linear gradient between the binding buffer and elution buffer A (20 mM Tris, pH 7.4, 500 mM NaCl, and 500 mM imidazole), followed by another 30 ml of 100% elution buffer A. The target protein was further purified by a 120 ml Superdex 200 (GE Healthcare) column with elution buffer B (20 mM Tris, pH 7.4, 200 mM NaCl). The N-terminal His-sumo-tagged protease was digested with sumo protease to remove the tag and then loaded onto a 120 ml Superdex 200 column. The proteins were again eluted with elution buffer B and concentrated after SDS-PAGE analysis. All of the purification procedures were performed at 4 °C to avoid unexpected degradation.
4.2. Crystallization and structure determination
PEDV 3CLpro was crystallized via the sitting drop vapor diffusion method at 20 °C. The best crystals were produced under the following conditions: 0.1 M NaCl, 0.2 M Na2HPO4: citric acid (pH 4.2) and 5% (w/v) PEG 3350. PEDV 3CLpro variant (C144A) crystals were obtained under the same conditions. The 11-amino-acid peptide substrate (TSAVLQ↓SGFRK, that contains the N-terminal auto-cleavage site of SARS-CoV 3CLpro) was dissolved at a 20 mM in elution buffer B as a stock solution. For crystal soaking, the solution was diluted to 10 mM and then added to the variant (C144A) crystallization drop at an equal volume. The crystals were soaked for 14 h before data collection. The single crystals were first washed with 5%, 10%, 15%, and 30% ethylene glycol (v/v) as a cryoprotectant and then flash-frozen in liquid nitrogen. All data collection was performed at beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF) using a MAR 225 CCD detector (MAR Research). All of the obtained data sets were indexed, integrated and scaled using HKL-3000 (Otwinowski and Minor, 1997). The structure was solved by molecular replacement with PHASER (Mccoy et al., 2007) using the structure of human coronavirus 229E 3CLpro (PDB identifier 1P9S) as a starting model. The structure of the complex was determined via molecular replacement using the previously determined PEDV 3CLpro structure as the search model. Manual model building was performed using Coot (Emsley and Cowtan, 2004), and the structure was refined with Phenix (Adams et al., 2002). Refinement statistics are shown in Table 1. All of the structural figures were drawn using PyMOL (Schrödinger, 2006).
Table 1.
Data and model statistics.
| PEDV 3CLpro | Complex of PEDV 3CLprovariant (C144A) bound to a peptide | |
|---|---|---|
| Data collection | ||
| Space group | P 1 21 1 | P 1 21 1 |
| Cell parameter(a, b, c(Å)) | 56.64, 91.06, 57.98 | 56.68, 91.88, 58.01 |
| α, β, γ | 90.00°,100.25°, 90.00° | 90.00°, 100.24°, 90.00° |
| Wavelength | 0.97917 | 0.97917 |
| Resolution range (Å) | 27.87−1.65 | 31.78–2.40 |
| % Completeness | 99.7 (99.9) | 99.8(99.9) |
| Rmerge (last shell) | 0.055(0.480) | 0.103(0.487) |
| I/σ (last shell) | 23.25(3.42) | 18.10(4.55) |
| Redundancy (last shell) | 3.7(3.7) | 3.2(3.1) |
| Refinement | ||
| Resolution (Å) | 27.45–1.65 | 31.78–2.40 |
| Rwork/Rfree | 17.9/20.3 | 16.5/21.8 |
| No. reflections | 69,111 | 22,868 |
| No of protein atoms | 4514 | 4512 |
| No. of solvent atoms | 465 | 267 |
| No. of ions/ligands | 0 | 1 |
| r.m.s.d. | ||
| Bond length (Å) | 0.007 | 0.008 |
| Bond angle (Å) | 1.126 | 1.100 |
| B factor (Å2) | 25.0 | 29.37 |
| Protein | 23.54 | 28.91 |
| Water | 33.49 | 33.60 |
| Ligand | 45.09 | |
| Ramachandran plot: core, | 97.78%, 2.22%, | 96.45%, 3.21%, |
| allow, disallow | 0.00% | 0.34% |
Highest resolution values are written in parenthesis.
Rmerge=Σ Σ |Ii−<I>|/Σ Σ Ii; where is Ii the intensity measurement of reflection h and <I>is the average intensity from multiple observations.
Rwork=Σ ||Fo|−|Fc||/Σ |Fo|; where Fo and Fc are the observed and calculated structure factors respectively.
Rfree is equivalent to Rwork but where 5% of the measured reflections have been excluded from refinement and set aside for cross-validation.
4.3. FRET-based assays for enzymatic characteristics
Based on the N-terminal cleavage site of PEDV 3CLpro, we designed the peptide substrate Dabcyl-YNSTLQ↓AGLRKM-E-Edans (Nanjing GenScript Company). The two fluorophores formed a quenching pair and exhibited fluorescence resonance energy transfer (FRET) within the peptide (Matayoshi et al., 1990). The increase in fluorescence upon cleavage of the fluorogenic peptide substrate was monitored every minute for 1 h at 485 nm, with excitation at 340 nm, using a fluorescence plate reader (Kuo et al., 2004). The WT- Δ3aa-His protein was used in the assays. All reactions were performed in 20 mM HEPES, 50 mM NaCl, 0.4 mM EDTA, 30% glycerol and 4 mM DTT at pH 8.0 in a total volume of 100 μl. The enzyme concentration used in the FRET assay was 200 nM, and the substrate concentration was 0–200 μM. The initial rates were used to calculate the kinetic parameters by fitting with the Michaelis-Menten equation using GraphPad Prism5. For the variants, all of the assay conditions were identical, but the assays were performed at a fixed substrate concentration.
4.4. Size-exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) analysis
The residue-residue interactions at the dimer interface of PEDV 3CLpro are listed in Table 2. Four variants (R4A, S138A, R294A and Q295A) and an N-finger deletion variant (Δ8aa) were selected for oligomeric state analysis. All of the mutated proteins were stored in elution buffer B at −80 °C. A Superdex 75 10/300 GL column (GE Healthcare) was used for the analysis of the WT and protein variants. The column was equilibrated with elution buffer B (20 mM Tris, pH 7.4, and 200 mM NaCl), and 250 μg of protein was then loaded onto the column and eluted with elution buffer B. Equal volumes of size exclusion standards (Conalbumin, 75 kDa; carbonic anhydrase, 29 kDa; equine myoglobin, 17 kDa; ribonuclease A, 13.7 kDa; aprotinin, 6.5 kDa; vitamin B12, 1.35 kDa) (Bio-Rad and GE Healthcare) were used to calibrate the column. The 280 nm absorbance peaks were generated and overlaid using Bio-Rad NGC software.
To further confirm the oligomeric state and determine the effect of the variants on the monomer-dimer equilibrium of PEDV 3CLpro, sedimentation velocity experiments were conducted at 18 °C on the XL-A instrument (Beckman) at 50,000 rpm. Proteins were prepared at the concentration of approximately 0.9 mg/ml in elution buffer B. The sedimentation boundary was monitored every 3 min for a total of 110 scans using the absorbance optics of 280 nm. The software Sedfit was used to fit the data to a distribution of Lamm equation solutions c(s) model (Schuck, 2000).
Accession numbers
Coordinate and structure factors have been submitted to the PDB (accession numbers 4XFQ and 4ZUH).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31372440) and the Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (Program Nos. 2012RC008, 2013PY031 and 2662015JQ003). We thank Joe Pasquarella and Dr. Fang Li for discussion and the staff at the SSRF BL-17U beamline for assistance in data collection. Moreover, we also thank research associates at Center for Protein Research (CPR), Huazhong agricultural University, for technical support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2016.04.018.
Appendix A. Supplementary material
Supplementary material FIGURE S1. An abnormal conformation of H41. The catalytic dyad of the PEDV 3CLpro structure (magenta) superimposed with the complex structure of the variant (C144A) (green). The two H41 imidazole rings have different orientations. The substrate is shown as yellow sticks. In the complex structure, the H41 imidazole ring is parallel with the P2-L side chain.
.
References
- Adams M.J., Carstens E.B. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses. Arch. Virol. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Chuck C.P., Chong L.T., Chen C., Chow H.F., Wan D.C., Wong K.B. Profiling of substrate specificity of SARS-CoV 3CL. Plos One. 2010;5:e13197. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C.P., Chow H.F., Wan D.C., Wong K.B. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. Plos One. 2011;6:e27228. doi: 10.1371/journal.pone.0027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin E.A., Anbalagan S., Okda F., Batman R., Nelson E., Hause B.M. An inactivated vaccine made from a U.S. field isolate of porcine epidemic disease virus is immunogenic in pigs as demonstrated by a dose-titration. BMC Vet. Res. 2015;11:62. doi: 10.1186/s12917-015-0357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Lau S.K.P., Woo P.C.Y. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns”. J. Formos. Med. Assoc. 2013;112:372–381. doi: 10.1016/j.jfma.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang J., Hu T., Chen K., Jiang H., Shen X. Residues on the dimer interface of SARS coronavirus 3C-like protease: dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 2008;143:525–536. doi: 10.1093/jb/mvm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Family Coronaviridae. In: King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B., editors. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; Oxford: 2012. pp. 806–828. [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garwes D.J. Transmissible gastroenteritis. Vet. Rec. 1988;122:462–463. doi: 10.1136/vr.122.19.462. [DOI] [PubMed] [Google Scholar]
- Hegyi A., Friebe A., Gorbalenya A.E., Ziebuhr J. Mutational analysis of the active centre of coronavirus 3C-like proteases. J. Gen. Virol. 2002;83:581–593. doi: 10.1099/0022-1317-83-3-581. [DOI] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hsu W.-C., Chang H.-C., Chou C.-Y., Tsai P.-J., Lin P.-I., Chang G.-G. Critical assessment of important regions in the subunit association and catalytic action of the severe acute respiratory syndrome coronavirus main protease. J. Biol. Chem. 2005;280:22741–22748. doi: 10.1074/jbc.M502556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-J., Chi Y.-H., Hsu J.T.-A., Liang P.-H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004;318:862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Brown T.D. Characterisation and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Denison M.R. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology. 1997;230:335–342. doi: 10.1006/viro.1997.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Qi Y., Teng X., Yang Z., Wei P., Zhang C., Tan L., Zhou L., Liu Y., Lai L. Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase. J. Biol. Chem. 2010;285:28134–28140. doi: 10.1074/jbc.M109.095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Lu X., Denison M.R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianayagam N.J., Sunde M., Matthews J.M. The power of two: protein dimerization in biology. Trends Biochem. Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Matayoshi E.D., Wang G.T., Krafft G.A., Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- Mccoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needle D., Lountos G.T., Waugh D.S. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015;71:1102–1111. doi: 10.1107/S1399004715003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham J. Letter to the editor. Pig Farming, 1972, 10, 723.
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pasick J., Berhane Y., Ojkic D., Maxie G., Embury-Hyatt C., Swekla K., Handel K., Fairles J., Alexandersen S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound. Emerg. Dis. 2014;61:397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin. Exp. Vaccin Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82:4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, L.L.C., 2006. The PyMOL Molecular Graphics System, Version 0.99r1.
- Tan J.Z., George S., Kusov Y., Perbandt M., Anemuller S., Mesters J.R., Norder H., Coutard B., Lacroix C., Leyssen P., Neyts J., Hilgenfeld R. 3C Protease of enterovirus 68: structure-based design of michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 2013;87:4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tomar S., Johnston M.L., John St.S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. Ligand-induced dimerization of MERS coronavirus nsp5 protease (3CLpro): implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015 doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q.H., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.Y., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L.R., Shi Y.L., Zhang H., Gao L., Peng G.Q., Chen H.C., Li K., Xiao S.B. Porcine epidemic diarrhea virus 3C-Like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Fan K.Q., Chen H., Ma L., Huang C.K., Tan L., Xi D., Li C.M., Liu Y., Cao A.N., Lai L.H. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2006;339:865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69 doi: 10.1128/MMBR.69.4.635-664.2005. 635−+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J. Design of wide-spectrum inhibitors targeting coronavirus main proteases. Plos Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Siddell S.G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 1999;73:177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material FIGURE S1. An abnormal conformation of H41. The catalytic dyad of the PEDV 3CLpro structure (magenta) superimposed with the complex structure of the variant (C144A) (green). The two H41 imidazole rings have different orientations. The substrate is shown as yellow sticks. In the complex structure, the H41 imidazole ring is parallel with the P2-L side chain.