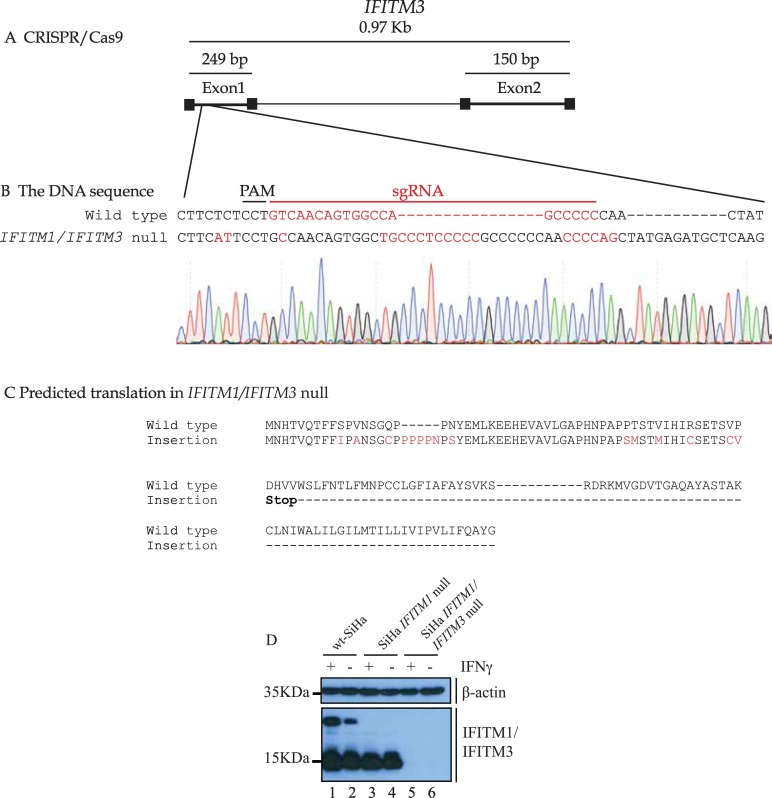
Fig. 3.
Validation of the IFITM3 guide RNA. (A). Gene structure of the IFITM3 gene highlighting its two coding exons and one intron. The genomic region surrounding the gRNA target site (with PAM site and homology region in red) for IFITM3 was amplified and subjected to DNA sequencing. The raw DNA sequencing reads in the IFITM3 gene are highlighted in (B). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The IFITM3 gene has two insertions that result in a poly-proline frame-shift creating a stop codon (C). (D). Immunoblotting was performed using Mab-MHK with lysates from SiHa cells (lanes 1 and 2); IFITM1 single null cells (lanes 3 and 4); and IFITM1/IFITM3 double null cells (lanes 5 and 6). The cells were either untreated or treated with IFNγ (100 ng/ml) for 24 h and processed for immunoblotting with either a β-actin antibody or Mab-MHK.