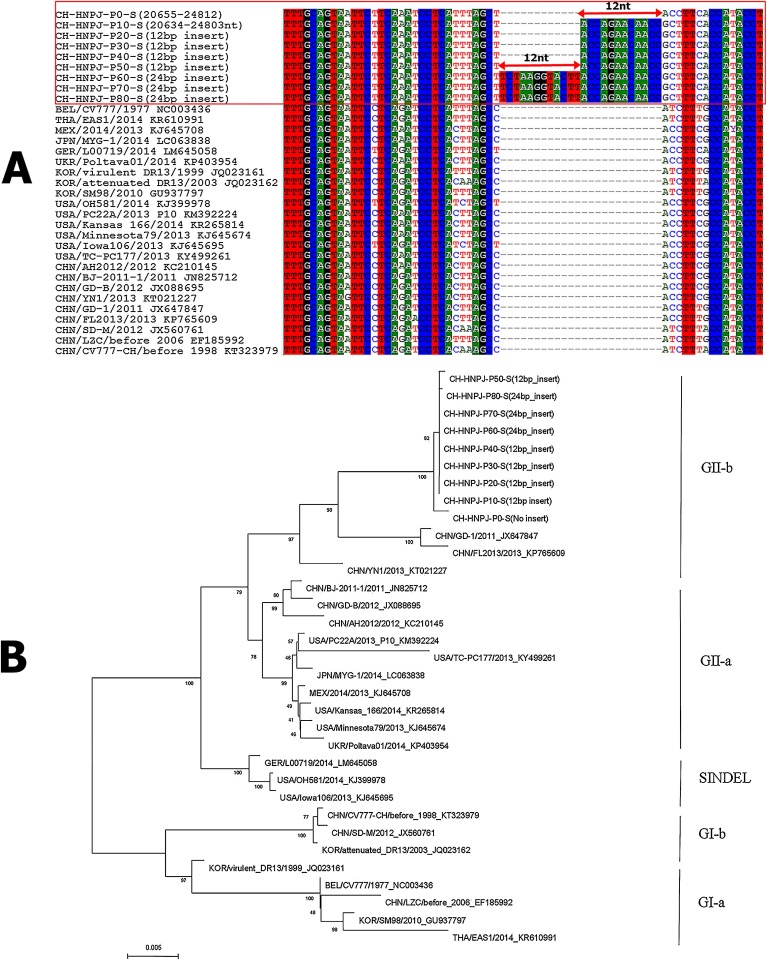
Fig. 4.
Nucleotide alignment and phylogenetic analysis of the S gene of the CH/HNPJ/2017 strain. (A) Alignments of the complete S gene of the parental (P0) and all cell culture-passaged CH/HNPJ/2017 strains with 10 generation intervals (P1, P10, P20, P30, P40, P50, P60, P70 and P80) and the reference sequence of the S gene of other PEDV strains are shown. The red solid box represents the CH/HNPJ/2017 sequence. The double-headed red arrows indicate inserted sequences. (B) Phylogenetic analysis based on the nucleotide sequences of the S gene of the parental (P0) and all cell culture-passaged CH/HNPJ/2017 strains with 10 generation intervals. The tree was constructed with the neighbor-joining method, and bootstrap values from 1000 resamplings are shown for each node. The country origin of the strains, years, GenBank accession numbers, genogroups and subgroups proposed in this study are shown (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).